The strain-encoded relationship between PrP replication, stability and processing in neurons is predictive of the incubation period of disease
- PMID: 21437239
- PMCID: PMC3060105
- DOI: 10.1371/journal.ppat.1001317
The strain-encoded relationship between PrP replication, stability and processing in neurons is predictive of the incubation period of disease
Abstract
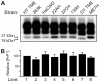
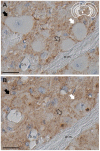
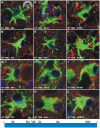
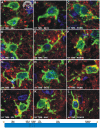
Prion strains are characterized by differences in the outcome of disease, most notably incubation period and neuropathological features. While it is established that the disease specific isoform of the prion protein, PrP(Sc), is an essential component of the infectious agent, the strain-specific relationship between PrP(Sc) properties and the biological features of the resulting disease is not clear. To investigate this relationship, we examined the amplification efficiency and conformational stability of PrP(Sc) from eight hamster-adapted prion strains and compared it to the resulting incubation period of disease and processing of PrP(Sc) in neurons and glia. We found that short incubation period strains were characterized by more efficient PrP(Sc) amplification and higher PrP(Sc) conformational stabilities compared to long incubation period strains. In the CNS, the short incubation period strains were characterized by the accumulation of N-terminally truncated PrP(Sc) in the soma of neurons, astrocytes and microglia in contrast to long incubation period strains where PrP(Sc) did not accumulate to detectable levels in the soma of neurons but was detected in glia similar to short incubation period strains. These results are inconsistent with the hypothesis that a decrease in conformational stability results in a corresponding increase in replication efficiency and suggest that glia mediated neurodegeneration results in longer survival times compared to direct replication of PrP(Sc) in neurons.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Flow Cytometric Detection of PrPSc in Neurons and Glial Cells from Prion-Infected Mouse Brains.J Virol. 2017 Dec 14;92(1):e01457-17. doi: 10.1128/JVI.01457-17. Print 2018 Jan 1. J Virol. 2017. PMID: 29046463 Free PMC article.
-
Incongruity between Prion Conversion and Incubation Period following Coinfection.J Virol. 2016 May 27;90(12):5715-23. doi: 10.1128/JVI.00409-16. Print 2016 Jun 15. J Virol. 2016. PMID: 27053546 Free PMC article.
-
Prion strain targeting independent of strain-specific neuronal tropism.J Virol. 2009 Jan;83(1):81-7. doi: 10.1128/JVI.01745-08. Epub 2008 Oct 29. J Virol. 2009. PMID: 18971281 Free PMC article.
-
Prion encephalopathies of animals and humans.Dev Biol Stand. 1993;80:31-44. Dev Biol Stand. 1993. PMID: 8270114 Review.
-
Neuropathology of prion diseases.Br Med Bull. 2003;66:121-30. doi: 10.1093/bmb/66.1.121. Br Med Bull. 2003. PMID: 14522854 Review.
Cited by
-
Changes in prion replication environment cause prion strain mutation.FASEB J. 2013 Sep;27(9):3702-10. doi: 10.1096/fj.13-230466. Epub 2013 May 31. FASEB J. 2013. PMID: 23729586 Free PMC article.
-
The role of prion strain diversity in the development of successful therapeutic treatments.Prog Mol Biol Transl Sci. 2020;175:77-119. doi: 10.1016/bs.pmbts.2020.07.001. Epub 2020 Aug 28. Prog Mol Biol Transl Sci. 2020. PMID: 32958242 Free PMC article. Review.
-
Stabilization of a prion strain of synthetic origin requires multiple serial passages.J Biol Chem. 2012 Aug 31;287(36):30205-14. doi: 10.1074/jbc.M112.392985. Epub 2012 Jul 17. J Biol Chem. 2012. PMID: 22807452 Free PMC article.
-
Assessment of strain-specific PrP(Sc) elongation rates revealed a transformation of PrP(Sc) properties during protein misfolding cyclic amplification.PLoS One. 2012;7(7):e41210. doi: 10.1371/journal.pone.0041210. Epub 2012 Jul 17. PLoS One. 2012. PMID: 22815972 Free PMC article.
-
Flow Cytometric Detection of PrPSc in Neurons and Glial Cells from Prion-Infected Mouse Brains.J Virol. 2017 Dec 14;92(1):e01457-17. doi: 10.1128/JVI.01457-17. Print 2018 Jan 1. J Virol. 2017. PMID: 29046463 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

