Genomes of the most dangerous epidemic bacteria have a virulence repertoire characterized by fewer genes but more toxin-antitoxin modules
- PMID: 21437250
- PMCID: PMC3060909
- DOI: 10.1371/journal.pone.0017962
Genomes of the most dangerous epidemic bacteria have a virulence repertoire characterized by fewer genes but more toxin-antitoxin modules
Abstract
Background: We conducted a comparative genomic study based on a neutral approach to identify genome specificities associated with the virulence capacity of pathogenic bacteria. We also determined whether virulence is dictated by rules, or if it is the result of individual evolutionary histories. We systematically compared the genomes of the 12 most dangerous pandemic bacteria for humans ("bad bugs") to their closest non-epidemic related species ("controls").
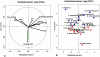
Methodology/principal findings: We found several significantly different features in the "bad bugs", one of which was a smaller genome that likely resulted from a degraded recombination and repair system. The 10 Cluster of Orthologous Group (COG) functional categories revealed a significantly smaller number of genes in the "bad bugs", which lacked mostly transcription, signal transduction mechanisms, cell motility, energy production and conversion, and metabolic and regulatory functions. A few genes were identified as virulence factors, including secretion system proteins. Five "bad bugs" showed a greater number of poly (A) tails compared to the controls, whereas an elevated number of poly (A) tails was found to be strongly correlated to a low GC% content. The "bad bugs" had fewer tandem repeat sequences compared to controls. Moreover, the results obtained from a principal component analysis (PCA) showed that the "bad bugs" had surprisingly more toxin-antitoxin modules than did the controls.
Conclusions/significance: We conclude that pathogenic capacity is not the result of "virulence factors" but is the outcome of a virulent gene repertoire resulting from reduced genome repertoires. Toxin-antitoxin systems could participate in the virulence repertoire, but they may have developed independently of selfish evolution.
Conflict of interest statement
Figures

Similar articles
-
Comparative genomics evidence that only protein toxins are tagging bad bugs.Front Cell Infect Microbiol. 2011 Oct 25;1:7. doi: 10.3389/fcimb.2011.00007. eCollection 2011. Front Cell Infect Microbiol. 2011. PMID: 22919573 Free PMC article.
-
Genomics of epidemic pathogens.Clin Microbiol Infect. 2012 Mar;18(3):213-7. doi: 10.1111/j.1469-0691.2012.03781.x. Clin Microbiol Infect. 2012. PMID: 22369153 Review.
-
Postgenomic analysis of bacterial pathogens repertoire reveals genome reduction rather than virulence factors.Brief Funct Genomics. 2013 Jul;12(4):291-304. doi: 10.1093/bfgp/elt015. Epub 2013 Jun 29. Brief Funct Genomics. 2013. PMID: 23814139
-
Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes.Biol Direct. 2009 Jun 3;4:19. doi: 10.1186/1745-6150-4-19. Biol Direct. 2009. PMID: 19493340 Free PMC article.
-
Novel toxins from type II toxin-antitoxin systems with acetyltransferase activity.Plasmid. 2017 Sep;93:30-35. doi: 10.1016/j.plasmid.2017.08.005. Epub 2017 Sep 20. Plasmid. 2017. PMID: 28941941 Review.
Cited by
-
Deciphering genomic virulence traits of a Staphylococcus epidermidis strain causing native-valve endocarditis.J Clin Microbiol. 2013 May;51(5):1617-21. doi: 10.1128/JCM.02820-12. Epub 2013 Jan 30. J Clin Microbiol. 2013. PMID: 23363834 Free PMC article.
-
Epidemiology, Clinical Aspects, Laboratory Diagnosis and Treatment of Rickettsial Diseases in the Mediterranean Area During COVID-19 Pandemic: A Review of the Literature.Mediterr J Hematol Infect Dis. 2020 Sep 1;12(1):e2020056. doi: 10.4084/MJHID.2020.056. eCollection 2020. Mediterr J Hematol Infect Dis. 2020. PMID: 32952967 Free PMC article. Review.
-
Multiple toxin-antitoxin systems in Mycobacterium tuberculosis.Toxins (Basel). 2014 Mar 6;6(3):1002-20. doi: 10.3390/toxins6031002. Toxins (Basel). 2014. PMID: 24662523 Free PMC article. Review.
-
Physiologic Stresses Reveal a Salmonella Persister State and TA Family Toxins Modulate Tolerance to These Stresses.PLoS One. 2015 Dec 3;10(12):e0141343. doi: 10.1371/journal.pone.0141343. eCollection 2015. PLoS One. 2015. PMID: 26633172 Free PMC article.
-
The VapBC-4 Characterization Indicates It Is a Bona Fide Toxin-Antitoxin Module of Leptospira interrogans: Initial Evidence for a Role in Bacterial Adaptation.Microorganisms. 2025 Apr 11;13(4):879. doi: 10.3390/microorganisms13040879. Microorganisms. 2025. PMID: 40284715 Free PMC article.
References
-
- Wu H-J, Wang H-J A, Jennings MP. Discovery of virulence factors of pathogenic bacteria. Curr Opin Chem Biol. 2008;12:1–9. - PubMed
-
- MC ten Bokum A, Movahedzadeh F, Frita R, Bancroft JG, Stoker GN. The case of hypervirulence through gene deletion in Mycobacterium tuberculosis. Trends Microbiol. 2008;16(9):436–441. - PubMed
-
- Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. - PubMed
-
- Sakharkar RK, Dhar KP, Chow TKV. Genome reduction in prokaryotic obligatory intracellular parasites of humans: a comparative analysis. Int J Syst Evol Microbiol. 2004;54:1937–1941. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

