Viral genome segmentation can result from a trade-off between genetic content and particle stability
- PMID: 21437265
- PMCID: PMC3060069
- DOI: 10.1371/journal.pgen.1001344
Viral genome segmentation can result from a trade-off between genetic content and particle stability
Abstract
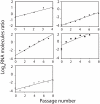
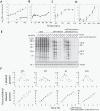
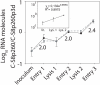
The evolutionary benefit of viral genome segmentation is a classical, yet unsolved question in evolutionary biology and RNA genetics. Theoretical studies anticipated that replication of shorter RNA segments could provide a replicative advantage over standard size genomes. However, this question has remained elusive to experimentalists because of the lack of a proper viral model system. Here we present a study with a stable segmented bipartite RNA virus and its ancestor non-segmented counterpart, in an identical genomic nucleotide sequence context. Results of RNA replication, protein expression, competition experiments, and inactivation of infectious particles point to a non-replicative trait, the particle stability, as the main driver of fitness gain of segmented genomes. Accordingly, measurements of the volume occupation of the genome inside viral capsids indicate that packaging shorter genomes involves a relaxation of the packaging density that is energetically favourable. The empirical observations are used to design a computational model that predicts the existence of a critical multiplicity of infection for domination of segmented over standard types. Our experiments suggest that viral segmented genomes may have arisen as a molecular solution for the trade-off between genome length and particle stability. Genome segmentation allows maximizing the genetic content without the detrimental effect in stability derived from incresing genome length.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


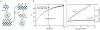
References
-
- Garcia-Arriaza J, Ojosnegros S, Davila M, Domingo E, Escarmis C. Dynamics of mutation and recombination in a replicating population of complementing, defective viral genomes. J Mol Biol. 2006;360:558–572. - PubMed
-
- Nee S. The evolution of multicompartmental genomes in viruses. J Mol Evol. 1987;25:277–281. - PubMed
-
- Szathmary E. Natural selection and dynamical coexistence of defective and complementing virus segments. J Theor Biol. 1992;157:383–406. - PubMed
-
- Chao L. Evolution of sex in RNA viruses. J Theor Biol. 1988;133:99–112. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

