Origin-dependent inverted-repeat amplification: a replication-based model for generating palindromic amplicons
- PMID: 21437266
- PMCID: PMC3060070
- DOI: 10.1371/journal.pgen.1002016
Origin-dependent inverted-repeat amplification: a replication-based model for generating palindromic amplicons
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
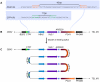
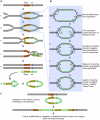

References
-
- Huang T, Campbell JL. Amplification of a circular episome carrying an inverted repeat of the DFR1 locus and adjacent autonomously replicating sequence element of Saccharomyces cerevisiae. J Biol Chem. 1995;270:9607–9614. - PubMed
-
- Vialard F, Mignon-Ravix C, Parain D, Depetris D, Portnoi MF, et al. Mechanism of intrachromosomal triplications 15q11-q13: a new clinical report. Am J Med Genet A. 2003;118A:229–234. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

