Making connections between ultrafast protein folding kinetics and molecular dynamics simulations
- PMID: 21441105
- PMCID: PMC3076883
- DOI: 10.1073/pnas.1019552108
Making connections between ultrafast protein folding kinetics and molecular dynamics simulations
Abstract
Determining the rate of forming the truly folded conformation of ultrafast folding proteins is an important issue for both experiments and simulations. The double-norleucine mutant of the 35-residue villin subdomain is the focus of recent computer simulations with atomistic molecular dynamics because it is currently the fastest folding protein. The folding kinetics of this protein have been measured in laser temperature-jump experiments using tryptophan fluorescence as a probe of overall folding. The conclusion from the simulations, however, is that the rate determined by fluorescence is significantly larger than the rate of overall folding. We have therefore employed an independent experimental method to determine the folding rate. The decay of the tryptophan triplet-state in photoselection experiments was used to monitor the change in the unfolded population for a sequence of the villin subdomain with one amino acid difference from that of the laser temperature-jump experiments, but with almost identical equilibrium properties. Folding times obtained in a two-state analysis of the results from the two methods at denaturant concentrations varying from 1.5-6.0 M guanidinium chloride are in excellent agreement, with an average difference of only 20%. Polynomial extrapolation of all the data to zero denaturant yields a folding time of 220 (+100,-70) ns at 283 K, suggesting that under these conditions the barrier between folded and unfolded states has effectively disappeared--the so-called "downhill scenario."
Conflict of interest statement
The authors declare no conflict of interest.
Figures

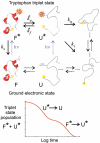
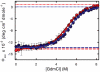
 ), the decay of the triplet-state population monitored by triplet–triplet optical absorption is biphasic, with the fast phase corresponding to the triplet-quenching rate in the unfolded state (kq), the slow phase to the unfolding rate (
), the decay of the triplet-state population monitored by triplet–triplet optical absorption is biphasic, with the fast phase corresponding to the triplet-quenching rate in the unfolded state (kq), the slow phase to the unfolding rate ( ), and the two amplitudes for the fast and slow phases given by the equilibrium fractions of the unfolded and folded states:
), and the two amplitudes for the fast and slow phases given by the equilibrium fractions of the unfolded and folded states:  and
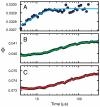
and  , respectively (, –36, 38). In the more realistic situation that we encounter with the double-norleucine mutant, Cys-HP35(Nle24,Nle29), where the timescales are not so clearly separable, the relaxation rates and amplitudes become complex functions of the four rate coefficients, which can be obtained by fitting the data with the solution of a simple differential equation model (see Eqs. 1–5).
, respectively (, –36, 38). In the more realistic situation that we encounter with the double-norleucine mutant, Cys-HP35(Nle24,Nle29), where the timescales are not so clearly separable, the relaxation rates and amplitudes become complex functions of the four rate coefficients, which can be obtained by fitting the data with the solution of a simple differential equation model (see Eqs. 1–5).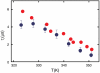


Similar articles
-
Evidence of multiple folding pathways for the villin headpiece subdomain.J Phys Chem B. 2011 Nov 3;115(43):12632-7. doi: 10.1021/jp206238y. Epub 2011 Oct 12. J Phys Chem B. 2011. PMID: 21923150
-
High-resolution x-ray crystal structures of the villin headpiece subdomain, an ultrafast folding protein.Proc Natl Acad Sci U S A. 2005 May 24;102(21):7517-22. doi: 10.1073/pnas.0502495102. Epub 2005 May 13. Proc Natl Acad Sci U S A. 2005. PMID: 15894611 Free PMC article.
-
Experimental tests of villin subdomain folding simulations.J Mol Biol. 2003 Jun 13;329(4):625-30. doi: 10.1016/s0022-2836(03)00519-9. J Mol Biol. 2003. PMID: 12787664
-
Sub-microsecond protein folding.J Mol Biol. 2006 Jun 9;359(3):546-53. doi: 10.1016/j.jmb.2006.03.034. Epub 2006 Mar 31. J Mol Biol. 2006. PMID: 16643946
-
Ultrafast and downhill protein folding.Curr Opin Struct Biol. 2007 Feb;17(1):38-47. doi: 10.1016/j.sbi.2007.01.001. Epub 2007 Jan 12. Curr Opin Struct Biol. 2007. PMID: 17223539 Review.
Cited by
-
Measuring ultrafast protein folding rates from photon-by-photon analysis of single molecule fluorescence trajectories.Chem Phys. 2013 Aug 30;422:229-237. doi: 10.1016/j.chemphys.2012.08.005. Chem Phys. 2013. PMID: 24443626 Free PMC article.
-
A quantitative connection of experimental and simulated folding landscapes by vibrational spectroscopy.Chem Sci. 2018 Oct 3;9(48):9002-9011. doi: 10.1039/c8sc03786h. eCollection 2018 Dec 28. Chem Sci. 2018. PMID: 30647892 Free PMC article.
-
Minimalist probes for studying protein dynamics: thioamide quenching of selectively excitable fluorescent amino acids.J Am Chem Soc. 2012 Apr 11;134(14):6088-91. doi: 10.1021/ja3005094. Epub 2012 Apr 3. J Am Chem Soc. 2012. PMID: 22471784 Free PMC article.
-
Quantitative Analysis of Protein Unfolded State Energetics: Experimental and Computational Studies Demonstrate That Non-Native Side-Chain Interactions Stabilize Local Native Backbone Structure.J Phys Chem B. 2021 Apr 8;125(13):3269-3277. doi: 10.1021/acs.jpcb.0c08922. Epub 2021 Mar 29. J Phys Chem B. 2021. PMID: 33779182 Free PMC article.
-
Spectroscopic studies of protein folding: linear and nonlinear methods.Protein Sci. 2012 Feb;21(2):157-70. doi: 10.1002/pro.2006. Epub 2011 Dec 28. Protein Sci. 2012. PMID: 22109973 Free PMC article. Review.
References
-
- Williams S, et al. Fast events in protein folding: Helix melting and formation in a small peptide. Biochemistry. 1996;35:691–697. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

