Corynebacterium glutamicum tailored for efficient isobutanol production
- PMID: 21441331
- PMCID: PMC3126470
- DOI: 10.1128/AEM.02972-10
Corynebacterium glutamicum tailored for efficient isobutanol production
Abstract
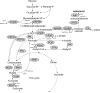
We recently engineered Corynebacterium glutamicum for aerobic production of 2-ketoisovalerate by inactivation of the pyruvate dehydrogenase complex, pyruvate:quinone oxidoreductase, transaminase B, and additional overexpression of the ilvBNCD genes, encoding acetohydroxyacid synthase, acetohydroxyacid isomeroreductase, and dihydroxyacid dehydratase. Based on this strain, we engineered C. glutamicum for the production of isobutanol from glucose under oxygen deprivation conditions by inactivation of l-lactate and malate dehydrogenases, implementation of ketoacid decarboxylase from Lactococcus lactis, alcohol dehydrogenase 2 (ADH2) from Saccharomyces cerevisiae, and expression of the pntAB transhydrogenase genes from Escherichia coli. The resulting strain produced isobutanol with a substrate-specific yield (Y(P/S)) of 0.60 ± 0.02 mol per mol of glucose. Interestingly, a chromosomally encoded alcohol dehydrogenase rather than the plasmid-encoded ADH2 from S. cerevisiae was involved in isobutanol formation with C. glutamicum, and overexpression of the corresponding adhA gene increased the Y(P/S) to 0.77 ± 0.01 mol of isobutanol per mol of glucose. Inactivation of the malic enzyme significantly reduced the Y(P/S), indicating that the metabolic cycle consisting of pyruvate and/or phosphoenolpyruvate carboxylase, malate dehydrogenase, and malic enzyme is responsible for the conversion of NADH + H+ to NADPH + H+. In fed-batch fermentations with an aerobic growth phase and an oxygen-depleted production phase, the most promising strain, C. glutamicum ΔaceE Δpqo ΔilvE ΔldhA Δmdh(pJC4ilvBNCD-pntAB)(pBB1kivd-adhA), produced about 175 mM isobutanol, with a volumetric productivity of 4.4 mM h⁻¹, and showed an overall Y(P/S) of about 0.48 mol per mol of glucose in the production phase.
Figures
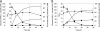
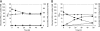

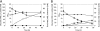

References
-
- Atsumi S., Hanai T., Liao J. C. 2008. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–89 - PubMed
-
- Becker J., et al. 2007. Metabolic flux engineering of l-lysine production in Corynebacterium glutamicum—over expression and modification of G6P dehydrogenase. J. Biotechnol. 132:99–109 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

