The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root
- PMID: 21441433
- PMCID: PMC3082253
- DOI: 10.1105/tpc.111.083352
The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root
Abstract
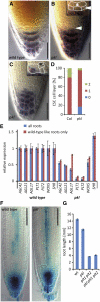
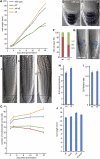
The chromatin modifying Polycomb group (PcG) and trithorax group (trxG) proteins are central regulators of cell identity that maintain a tightly controlled balance between cell proliferation and cell differentiation. The opposing activities of PcG and trxG proteins ensure the correct expression of specific transcriptional programs at defined developmental stages. Here, we report that the chromatin remodeling factor PICKLE (PKL) and the PcG protein CURLY LEAF (CLF) antagonistically determine root meristem activity. Whereas loss of PKL function caused a decrease in meristematic activity, loss of CLF function increased meristematic activity. Alterations of meristematic activity in pkl and clf mutants were not connected with changes in auxin concentration but correlated with decreased or increased expression of root stem cell and meristem marker genes, respectively. Root stem cell and meristem marker genes are modified by the PcG-mediated trimethylation of histone H3 on lysine 27 (H3K27me3). Decreased expression levels of root stem cell and meristem marker genes in pkl correlated with increased levels of H3K27me3, indicating that root meristem activity is largely controlled by the antagonistic activity of PcG proteins and PKL.
Figures






References
-
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.S., Amasino R., Scheres B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 - PubMed
-
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 - PubMed
-
- Bouazoune K., Brehm A. (2006). ATP-dependent chromatin remodeling complexes in Drosophila. Chromosome Res. 14: 433–449 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

