G-quadruplex formation at the 3' end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase
- PMID: 21441540
- PMCID: PMC3152327
- DOI: 10.1093/nar/gkr164
G-quadruplex formation at the 3' end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase
Abstract
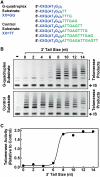
Telomere G-quadruplex is emerging as a promising anti-cancer target due to its inhibition to telomerase, an enzyme expressed in more than 85% tumors. Telomerase-mediated telomere extension and some other reactions require a free 3' telomere end in single-stranded form. G-quadruplex formation near the 3' end of telomere DNA can leave a 3' single-stranded tail of various sizes. How these terminal structures affect reactions at telomere end is not clear. In this work, we studied the 3' tail size-dependence of telomere extension by either telomerase or the alternative lengthening of telomere (ALT) mechanism as well as telomere G-quadruplex unwinding. We show that these reactions require a minimal tail of 8, 12 and 6 nt, respectively. Since we have shown that G-quadruplex tends to form at the farthest 3' distal end of telomere DNA leaving a tail of no more than 5 nt, these results imply that G-quadruplex formation may play a role in regulating reactions at the telomere ends and, as a result, serve as effective drug target for intervening telomere function.
Figures






References
-
- Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. - PubMed
-
- Chai W, Du Q, Shay JW, Wright WE. Human telomeres have different overhang sizes at leading versus lagging strands. Mol. Cell. 2006;21:427–435. - PubMed
-
- Olovnikov AM. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 1973;41:181–190. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

