Spliceosome structure and function
- PMID: 21441581
- PMCID: PMC3119917
- DOI: 10.1101/cshperspect.a003707
Spliceosome structure and function
Abstract
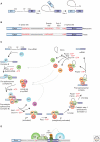
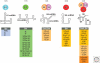
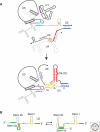
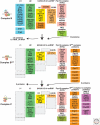
Pre-mRNA splicing is catalyzed by the spliceosome, a multimegadalton ribonucleoprotein (RNP) complex comprised of five snRNPs and numerous proteins. Intricate RNA-RNA and RNP networks, which serve to align the reactive groups of the pre-mRNA for catalysis, are formed and repeatedly rearranged during spliceosome assembly and catalysis. Both the conformation and composition of the spliceosome are highly dynamic, affording the splicing machinery its accuracy and flexibility, and these remarkable dynamics are largely conserved between yeast and metazoans. Because of its dynamic and complex nature, obtaining structural information about the spliceosome represents a major challenge. Electron microscopy has revealed the general morphology of several spliceosomal complexes and their snRNP subunits, and also the spatial arrangement of some of their components. X-ray and NMR studies have provided high resolution structure information about spliceosomal proteins alone or complexed with one or more binding partners. The extensive interplay of RNA and proteins in aligning the pre-mRNA's reactive groups, and the presence of both RNA and protein at the core of the splicing machinery, suggest that the spliceosome is an RNP enzyme. However, elucidation of the precise nature of the spliceosome's active site, awaits the generation of a high-resolution structure of its RNP core.
Figures



References
-
- Abelson J 2008. Is the spliceosome a ribonucleoprotein enzyme? Nat Struct Mol Biol 15: 1235–1237 - PubMed
-
- Ast G 2004. How did alternative splicing evolve? Nat Rev 5: 773–782 - PubMed
-
- Azubel M, Wolf SG, Sperling J, Sperling R 2004. Three-dimensional structure of the native spliceosome by cryo-electron microscopy. Mol Cell 15: 833–839 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
