Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice
- PMID: 21441929
- PMCID: PMC3084174
- DOI: 10.1038/ng.796
Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice
Abstract
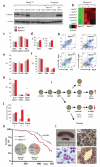
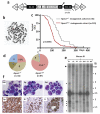
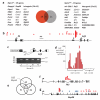
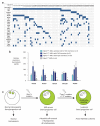
Acute myeloid leukemia (AML) is a molecularly diverse malignancy with a poor prognosis whose largest subgroup is characterized by somatic mutations in NPM1, which encodes nucleophosmin. These mutations, termed NPM1c, result in cytoplasmic dislocation of nucleophosmin and are associated with distinctive transcriptional signatures, yet their role in leukemogenesis remains obscure. Here we report that activation of a humanized Npm1c knock-in allele in mouse hemopoietic stem cells causes Hox gene overexpression, enhanced self renewal and expanded myelopoiesis. One third of mice developed delayed-onset AML, suggesting a requirement for cooperating mutations. We identified such mutations using a Sleeping Beauty transposon, which caused rapid-onset AML in 80% of mice with Npm1c, associated with mutually exclusive integrations in Csf2, Flt3 or Rasgrp1 in 55 of 70 leukemias. We also identified recurrent integrations in known and newly discovered leukemia genes including Nf1, Bach2, Dleu2 and Nup98. Our results provide new pathogenetic insights and identify possible therapeutic targets in NPM1c+ AML.
Figures

References
-
- Falini B, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. - PubMed
-
- Alcalay M, et al. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood. 2005;106:899–902. doi:2005-02-0560 [pii] 10.1182/blood-2005-02-0560. - PubMed
-
- Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. - PubMed
-
- Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi:nature03681 [pii] 10.1038/nature03681. - PubMed
-
- Okuwaki M. The structure and functions of NPM1/Nucleophsmin/B23, a multifunctional nucleolar acidic protein. J Biochem. 2008;143:441–448. doi:mvm222 [pii] 10.1093/jb/mvm222. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

