A lateral belt of cortical LGN and NuMA guides mitotic spindle movements and planar division in neuroepithelial cells
- PMID: 21444683
- PMCID: PMC3082188
- DOI: 10.1083/jcb.201101039
A lateral belt of cortical LGN and NuMA guides mitotic spindle movements and planar division in neuroepithelial cells
Abstract
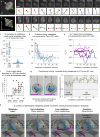
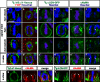
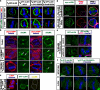
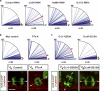
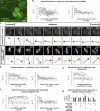
To maintain tissue architecture, epithelial cells divide in a planar fashion, perpendicular to their main polarity axis. As the centrosome resumes an apical localization in interphase, planar spindle orientation is reset at each cell cycle. We used three-dimensional live imaging of GFP-labeled centrosomes to investigate the dynamics of spindle orientation in chick neuroepithelial cells. The mitotic spindle displays stereotypic movements during metaphase, with an active phase of planar orientation and a subsequent phase of planar maintenance before anaphase. We describe the localization of the NuMA and LGN proteins in a belt at the lateral cell cortex during spindle orientation. Finally, we show that the complex formed of LGN, NuMA, and of cortically located Gαi subunits is necessary for spindle movements and regulates the dynamics of spindle orientation. The restricted localization of LGN and NuMA in the lateral belt is instructive for the planar alignment of the mitotic spindle, and required for its planar maintenance.
Figures
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

