A hierarchy of signals regulates entry of membrane proteins into the ciliary membrane domain in epithelial cells
- PMID: 21444686
- PMCID: PMC3082196
- DOI: 10.1083/jcb.201009001
A hierarchy of signals regulates entry of membrane proteins into the ciliary membrane domain in epithelial cells
Abstract
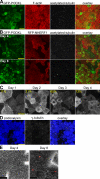
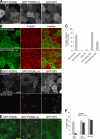
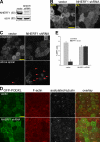
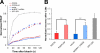
The membrane of the primary cilium is continuous with the plasma membrane but compositionally distinct. Although some membrane proteins concentrate in the cilium, others such as podocalyxin/gp135 are excluded. We found that exclusion reflects a saturable selective retention mechanism. Podocalyxin is immobilized by its PDZ interaction motif binding to NHERF1 and thereby to the apical actin network via ERM family members. The retention signal was dominant, autonomous, and transferable to membrane proteins not normally excluded from the cilium. The NHERF1-binding domains of cystic fibrosis transmembrane conductance regulator and Csk-binding protein were also found to act as transferable retention signals. Addition of a retention signal could inhibit the ciliary localization of proteins (e.g., Smoothened) containing signals that normally facilitate concentration in the ciliary membrane. Proteins without a retention signal (e.g., green fluorescent protein-glycosylphosphatidylinositol) were found in the cilium, suggesting entry was not impeded by a diffusion barrier or lipid microdomain. Thus, a hierarchy of interactions controls the composition of the ciliary membrane, including selective retention, selective inclusion, and passive diffusion.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

