Replication stress induces 53BP1-containing OPT domains in G1 cells
- PMID: 21444690
- PMCID: PMC3082192
- DOI: 10.1083/jcb.201011083
Replication stress induces 53BP1-containing OPT domains in G1 cells
Abstract
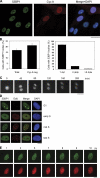
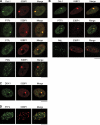
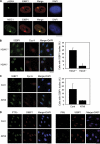
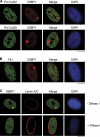
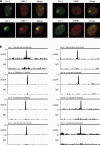
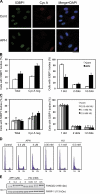
Chromosomal deletions and rearrangements in tumors are often associated with common fragile sites, which are specific genomic loci prone to gaps and breaks in metaphase chromosomes. Common fragile sites appear to arise through incomplete DNA replication because they are induced after partial replication inhibition by agents such as aphidicolin. Here, we show that in G1 cells, large nuclear bodies arise that contain p53 binding protein 1 (53BP1), phosphorylated H2AX (γH2AX), and mediator of DNA damage checkpoint 1 (MDC1), as well as components of previously characterized OPT (Oct-1, PTF, transcription) domains. Notably, we find that incubating cells with low aphidicolin doses increases the incidence and number of 53BP1-OPT domains in G1 cells, and by chromatin immunoprecipitation and massively parallel sequencing analysis of γH2AX, we demonstrate that OPT domains are enriched at common fragile sites. These findings invoke a model wherein incomplete DNA synthesis during S phase leads to a DNA damage response and formation of 53BP1-OPT domains in the subsequent G1.
Figures
References
-
- Bouwman P., Aly A., Escandell J.M., Pieterse M., Bartkova J., van der Gulden H., Hiddingh S., Thanasoula M., Kulkarni A., Yang Q., et al. 2010. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17:688–695 10.1038/nsmb.1831 - DOI - PMC - PubMed
-
- Bunting S.F., Callén E., Wong N., Chen H.T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 141:243–254 10.1016/j.cell.2010.03.012 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

