Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens
- PMID: 21444795
- PMCID: PMC3076887
- DOI: 10.1073/pnas.1019077108
Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens
Abstract
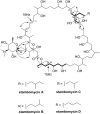
There is a constant need for new and improved drugs to combat infectious diseases, cancer, and other major life-threatening conditions. The recent development of genomics-guided approaches for novel natural product discovery has stimulated renewed interest in the search for natural product-based drugs. Genome sequence analysis of Streptomyces ambofaciens ATCC23877 has revealed numerous secondary metabolite biosynthetic gene clusters, including a giant type I modular polyketide synthase (PKS) gene cluster, which is composed of 25 genes (nine of which encode PKSs) and spans almost 150 kb, making it one of the largest polyketide biosynthetic gene clusters described to date. The metabolic product(s) of this gene cluster are unknown, and transcriptional analyses showed that it is not expressed under laboratory growth conditions. The constitutive expression of a regulatory gene within the cluster, encoding a protein that is similar to Large ATP binding of the LuxR (LAL) family proteins, triggered the expression of the biosynthetic genes. This led to the identification of four 51-membered glycosylated macrolides, named stambomycins A-D as metabolic products of the gene cluster. The structures of these compounds imply several interesting biosynthetic features, including incorporation of unusual extender units into the polyketide chain and in trans hydroxylation of the growing polyketide chain to provide the hydroxyl group for macrolide formation. Interestingly, the stambomycins possess promising antiproliferative activity against human cancer cell lines. Database searches identify genes encoding LAL regulators within numerous cryptic biosynthetic gene clusters in actinomycete genomes, suggesting that constitutive expression of such pathway-specific activators represents a powerful approach for novel bioactive natural product discovery.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Bérdy J. Bioactive microbial metabolites. J Antibiot (Tokyo) 2005;58:1–26. - PubMed
-
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. - PubMed
-
- Challis GL. Mining microbial genomes for new natural products and biosynthetic pathways. Microbiology. 2008;154:1555–1569. - PubMed
-
- Zerikly M, Challis GL. Strategies for the discovery of new natural products by genome mining. ChemBioChem. 2009;10:625–633. - PubMed
-
- Pinnert-Sindico S. Une nouvelle espèce de Streptomyces productrice d'antibiotiques: Streptomyces ambofaciens n. sp. caractères culturaux. Ann Inst Pasteur (Paris) 1954;87:702–707. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

