Gapped spectral dictionaries and their applications for database searches of tandem mass spectra
- PMID: 21444829
- PMCID: PMC3108828
- DOI: 10.1074/mcp.M110.002220
Gapped spectral dictionaries and their applications for database searches of tandem mass spectra
Abstract
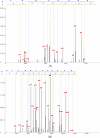
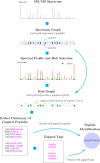
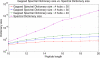
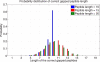
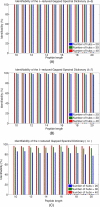
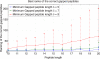
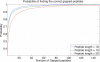
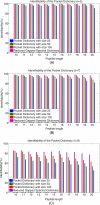
Generating all plausible de novo interpretations of a peptide tandem mass (MS/MS) spectrum (Spectral Dictionary) and quickly matching them against the database represent a recently emerged alternative approach to peptide identification. However, the sizes of the Spectral Dictionaries quickly grow with the peptide length making their generation impractical for long peptides. We introduce Gapped Spectral Dictionaries (all plausible de novo interpretations with gaps) that can be easily generated for any peptide length thus addressing the limitation of the Spectral Dictionary approach. We show that Gapped Spectral Dictionaries are small thus opening a possibility of using them to speed-up MS/MS searches. Our MS-Gapped-Dictionary algorithm (based on Gapped Spectral Dictionaries) enables proteogenomics applications (such as searches in the six-frame translation of the human genome) that are prohibitively time consuming with existing approaches. MS-Gapped-Dictionary generates gapped peptides that occupy a niche between accurate but short peptide sequence tags and long but inaccurate full length peptide reconstructions. We show that, contrary to conventional wisdom, some high-quality spectra do not have good peptide sequence tags and introduce gapped tags that have advantages over the conventional peptide sequence tags in MS/MS database searches.
Figures

Similar articles
-
Spectral dictionaries: Integrating de novo peptide sequencing with database search of tandem mass spectra.Mol Cell Proteomics. 2009 Jan;8(1):53-69. doi: 10.1074/mcp.M800103-MCP200. Epub 2008 Aug 14. Mol Cell Proteomics. 2009. PMID: 18703573 Free PMC article.
-
Spectral profiles, a novel representation of tandem mass spectra and their applications for de novo peptide sequencing and identification.Mol Cell Proteomics. 2009 Jun;8(6):1391-400. doi: 10.1074/mcp.M800535-MCP200. Epub 2009 Mar 2. Mol Cell Proteomics. 2009. PMID: 19254948 Free PMC article.
-
An efficient algorithm for the blocked pattern matching problem.Bioinformatics. 2015 Feb 15;31(4):532-8. doi: 10.1093/bioinformatics/btu678. Epub 2014 Oct 15. Bioinformatics. 2015. PMID: 25322837
-
The spectral networks paradigm in high throughput mass spectrometry.Mol Biosyst. 2012 Oct;8(10):2535-44. doi: 10.1039/c2mb25085c. Mol Biosyst. 2012. PMID: 22610447 Free PMC article. Review.
-
Algorithms for the de novo sequencing of peptides from tandem mass spectra.Expert Rev Proteomics. 2011 Oct;8(5):645-57. doi: 10.1586/epr.11.54. Expert Rev Proteomics. 2011. PMID: 21999834 Review.
Cited by
-
MS-GF+ makes progress towards a universal database search tool for proteomics.Nat Commun. 2014 Oct 31;5:5277. doi: 10.1038/ncomms6277. Nat Commun. 2014. PMID: 25358478 Free PMC article.
-
Speeding up tandem mass spectral identification using indexes.Bioinformatics. 2012 Jul 1;28(13):1692-7. doi: 10.1093/bioinformatics/bts244. Epub 2012 Apr 27. Bioinformatics. 2012. PMID: 22543365 Free PMC article.
-
De novo sequencing and homology searching.Mol Cell Proteomics. 2012 Feb;11(2):O111.014902. doi: 10.1074/mcp.O111.014902. Epub 2011 Nov 16. Mol Cell Proteomics. 2012. PMID: 22090170 Free PMC article.
-
Spectrum Identification using a Dynamic Bayesian Network Model of Tandem Mass Spectra.Uncertain Artif Intell. 2012 Aug;28:775-785. Uncertain Artif Intell. 2012. PMID: 25383048 Free PMC article.
-
The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search.Mol Cell Proteomics. 2010 Dec;9(12):2840-52. doi: 10.1074/mcp.M110.003731. Epub 2010 Sep 9. Mol Cell Proteomics. 2010. PMID: 20829449 Free PMC article.
References
-
- Ma B., Zhang K., Hendrie C., Liang C., Li M., Doherty-Kirby A., Lajoie G. (2003) PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 17, 2337–2342 - PubMed
-
- Frank A., Pevzner P. (2005) PepNovo: de novo peptide sequencing via probabilistic network modeling. Anal. Chem. 77, 964–973 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

