Molecular determinants of S100B oligomer formation
- PMID: 21445240
- PMCID: PMC3060798
- DOI: 10.1371/journal.pone.0014768
Molecular determinants of S100B oligomer formation
Abstract
Background: S100B is a dimeric protein that can form tetramers, hexamers and higher order oligomers. These forms have been suggested to play a role in RAGE activation.
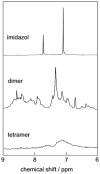
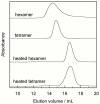
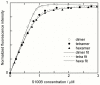
Methodology/principal findings: Oligomerization was found to require a low molecular weight trigger/cofactor and could not be detected for highly pure dimer, irrespective of handling. Imidazol was identified as a substance that can serve this role. Oligomerization is dependent on both the imidazol concentration and pH, with optima around 90 mM imidazol and pH 7, respectively. No oligomerization was observed above pH 8, thus the protonated form of imidazol is the active species in promoting assembly of dimers to higher species. However, disulfide bonds are not involved and the process is independent of redox potential. The process was also found to be independent of whether Ca(2+) is bound to the protein or not. Tetramers that are purified from dimers and imidazol by gel filtration are kinetically stable, but dissociate into dimers upon heating. Dimers do not revert to tetramer and higher oligomer unless imidazol is again added. Both tetramers and hexamers bind the target peptide from p53 with retained stoichiometry of one peptide per S100B monomer, and with high affinity (lgK = 7.3±0.2 and 7.2±0.2, respectively in 10 mM BisTris, 5 mM CaCl(2), pH 7.0), which is less than one order of magnitude reduced compared to dimer under the same buffer conditions.
Conclusion/significance: S100B oligomerization requires protonated imidazol as a trigger/cofactor. Oligomers are kinetically stable after imidazol is removed but revert back to dimer if heated. The results underscore the importance of kinetic versus thermodynamic control of S100B protein aggregation.
Conflict of interest statement
Figures


References
-
- Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. The international journal of biochemistry and cell biology. 2001;33:637–668. - PubMed
-
- Donato R. Intracellular and extracellular roles of S100 proteins. Microscopy Research and Technique. 2003;60:540–551. - PubMed
-
- Heizmann CW, Fritz G, Schäfer BW. S100 proteins: structure, functions and pathology. Front Biosci. 2002;7:d1356–1368. - PubMed
-
- Rothermundt M, Peters M, Prehn JHM, Arolt V. S100B in brain damage and neurodegeneration. Microscopy Research and Technique. 2003;60:614–632. - PubMed
-
- Schäfer BW, Heizmann CW. The S100 family of EF-hand calcium-binding proteins: functions and pathology. Trends Biochem Sci. 1996;21:134–40. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

