Selective constraints on amino acids estimated by a mechanistic codon substitution model with multiple nucleotide changes
- PMID: 21445250
- PMCID: PMC3060808
- DOI: 10.1371/journal.pone.0017244
Selective constraints on amino acids estimated by a mechanistic codon substitution model with multiple nucleotide changes
Abstract
Background: Empirical substitution matrices represent the average tendencies of substitutions over various protein families by sacrificing gene-level resolution. We develop a codon-based model, in which mutational tendencies of codon, a genetic code, and the strength of selective constraints against amino acid replacements can be tailored to a given gene. First, selective constraints averaged over proteins are estimated by maximizing the likelihood of each 1-PAM matrix of empirical amino acid (JTT, WAG, and LG) and codon (KHG) substitution matrices. Then, selective constraints specific to given proteins are approximated as a linear function of those estimated from the empirical substitution matrices.
Results: Akaike information criterion (AIC) values indicate that a model allowing multiple nucleotide changes fits the empirical substitution matrices significantly better. Also, the ML estimates of transition-transversion bias obtained from these empirical matrices are not so large as previously estimated. The selective constraints are characteristic of proteins rather than species. However, their relative strengths among amino acid pairs can be approximated not to depend very much on protein families but amino acid pairs, because the present model, in which selective constraints are approximated to be a linear function of those estimated from the JTT/WAG/LG/KHG matrices, can provide a good fit to other empirical substitution matrices including cpREV for chloroplast proteins and mtREV for vertebrate mitochondrial proteins.
Conclusions/significance: The present codon-based model with the ML estimates of selective constraints and with adjustable mutation rates of nucleotide would be useful as a simple substitution model in ML and Bayesian inferences of molecular phylogenetic trees, and enables us to obtain biologically meaningful information at both nucleotide and amino acid levels from codon and protein sequences.
Conflict of interest statement
Figures

 of the log-odds matrices of (A) the ML-87 and (B) the ML-91 models fitted to the 1-PAM JTT matrix is plotted against the log-odds log-
of the log-odds matrices of (A) the ML-87 and (B) the ML-91 models fitted to the 1-PAM JTT matrix is plotted against the log-odds log- calculated from JTT. Plus, circle, and cross marks show the log-odds values for the types of substitutions requiring single, double and triple nucleotide changes, respectively. The dotted line in each figure shows the line of equal values between the ordinate and the abscissa.
calculated from JTT. Plus, circle, and cross marks show the log-odds values for the types of substitutions requiring single, double and triple nucleotide changes, respectively. The dotted line in each figure shows the line of equal values between the ordinate and the abscissa.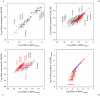
 of the log-odds matrix corresponding to (A) single, (B) double, and (C) triple nucleotide changes in the ML-200 model fitted to the 1-PAM KHG codon substitution matrix is plotted against the log-odds log-
of the log-odds matrix corresponding to (A) single, (B) double, and (C) triple nucleotide changes in the ML-200 model fitted to the 1-PAM KHG codon substitution matrix is plotted against the log-odds log- calculated from KHG. In (D), codon log-exchangeabilities of the 1-PAM KHG codon substitution matrix corresponding to triple nucleotide changes are plotted against the log-odds log-
calculated from KHG. In (D), codon log-exchangeabilities of the 1-PAM KHG codon substitution matrix corresponding to triple nucleotide changes are plotted against the log-odds log- calculated from KHG. The log-exchangeability of the 1-PAM KHG is defined as
calculated from KHG. The log-exchangeability of the 1-PAM KHG is defined as  . Upper triangle, plus, circle, and cross marks show the log-odds values for synonymous pairs and one-, two-, and three-step amino acid pairs, respectively. Log-exchangeabilities for the codon pairs whose instantaneous rates are estimated to be
. Upper triangle, plus, circle, and cross marks show the log-odds values for synonymous pairs and one-, two-, and three-step amino acid pairs, respectively. Log-exchangeabilities for the codon pairs whose instantaneous rates are estimated to be  in KHG are shown to be about
in KHG are shown to be about  in this figure. The dotted line in each figure shows the line of equal values between the ordinate and the abscissa.
in this figure. The dotted line in each figure shows the line of equal values between the ordinate and the abscissa.
 in the ML-91+ model fitted to the 1-PAM JTT amino acid substitution matrix and (B)
in the ML-91+ model fitted to the 1-PAM JTT amino acid substitution matrix and (B)  in the ML-200 model fitted to the 1-PAM KHG codon substitution matrix, for each amino acid pair is plotted against the mean energy increment due to an amino acid substitution, (
in the ML-200 model fitted to the 1-PAM KHG codon substitution matrix, for each amino acid pair is plotted against the mean energy increment due to an amino acid substitution, ( ) defined by Eqs. S1-4, S1-5, and S1-6 in Text S1. In (A), the estimates
) defined by Eqs. S1-4, S1-5, and S1-6 in Text S1. In (A), the estimates  for the least exchangeable class of multi-step amino acid pairs are not shown. Plus, circle, and cross marks show the values for one-, two-, and three-step amino acid pairs, respectively.
for the least exchangeable class of multi-step amino acid pairs are not shown. Plus, circle, and cross marks show the values for one-, two-, and three-step amino acid pairs, respectively.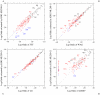
 of the log-odds matrices of the KHG-ML200-11 model fitted to the 1-PAM matrices of (A) JTT, (B) WAG, (C) LG, and (D) mtREV is plotted against the log-odds log-
of the log-odds matrices of the KHG-ML200-11 model fitted to the 1-PAM matrices of (A) JTT, (B) WAG, (C) LG, and (D) mtREV is plotted against the log-odds log- calculated from the corresponding empirical substitution matrices. Plus, circle, and cross marks show the log-odds values for one-, two-, and three-step amino acid pairs, respectively. The dotted line in each figure shows the line of equal values between the ordinate and the abscissa. The log-odds elements of mtREV whose values are smaller than about
calculated from the corresponding empirical substitution matrices. Plus, circle, and cross marks show the log-odds values for one-, two-, and three-step amino acid pairs, respectively. The dotted line in each figure shows the line of equal values between the ordinate and the abscissa. The log-odds elements of mtREV whose values are smaller than about  are all assumed to be
are all assumed to be  ; see the original paper .
; see the original paper .Similar articles
-
Advantages of a mechanistic codon substitution model for evolutionary analysis of protein-coding sequences.PLoS One. 2011;6(12):e28892. doi: 10.1371/journal.pone.0028892. Epub 2011 Dec 29. PLoS One. 2011. PMID: 22220197 Free PMC article.
-
Next-generation development and application of codon model in evolution.Front Genet. 2023 Jan 27;14:1091575. doi: 10.3389/fgene.2023.1091575. eCollection 2023. Front Genet. 2023. PMID: 36777719 Free PMC article. Review.
-
Superiority of a mechanistic codon substitution model even for protein sequences in phylogenetic analysis.BMC Evol Biol. 2013 Nov 21;13:257. doi: 10.1186/1471-2148-13-257. BMC Evol Biol. 2013. PMID: 24256155 Free PMC article.
-
A thermodynamic model of protein structure evolution explains empirical amino acid substitution matrices.Protein Sci. 2021 Oct;30(10):2057-2068. doi: 10.1002/pro.4155. Epub 2021 Jul 30. Protein Sci. 2021. PMID: 34218472 Free PMC article.
-
Models of amino acid substitution and applications to mitochondrial protein evolution.Mol Biol Evol. 1998 Dec;15(12):1600-11. doi: 10.1093/oxfordjournals.molbev.a025888. Mol Biol Evol. 1998. PMID: 9866196
Cited by
-
Combining extracellular matrix proteome and phosphoproteome of chickpea and meta-analysis reveal novel proteoforms and evolutionary significance of clade-specific wall-associated events in plant.Plant Direct. 2024 Mar 18;8(3):e572. doi: 10.1002/pld3.572. eCollection 2024 Mar. Plant Direct. 2024. PMID: 38500675 Free PMC article.
-
Advantages of a mechanistic codon substitution model for evolutionary analysis of protein-coding sequences.PLoS One. 2011;6(12):e28892. doi: 10.1371/journal.pone.0028892. Epub 2011 Dec 29. PLoS One. 2011. PMID: 22220197 Free PMC article.
-
Next-generation development and application of codon model in evolution.Front Genet. 2023 Jan 27;14:1091575. doi: 10.3389/fgene.2023.1091575. eCollection 2023. Front Genet. 2023. PMID: 36777719 Free PMC article. Review.
-
Prediction of contact residue pairs based on co-substitution between sites in protein structures.PLoS One. 2013;8(1):e54252. doi: 10.1371/journal.pone.0054252. Epub 2013 Jan 16. PLoS One. 2013. PMID: 23342110 Free PMC article.
-
Prevalence of multinucleotide replacements in evolution of primates and Drosophila.Mol Biol Evol. 2013 Jun;30(6):1315-25. doi: 10.1093/molbev/mst036. Epub 2013 Feb 27. Mol Biol Evol. 2013. PMID: 23447710 Free PMC article.
References
-
- Kimura M. A simple model for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. - PubMed
-
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial dna. J Mol Evol. 1985;22:160–174. - PubMed
-
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial dna in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. - PubMed
-
- Dayhoff MO, Schwartz RM, Orcutt BC. A model of evolutionary change in proteins. In: Dayhoff MO, editor. Atlas of protein sequence and structure. Washington D.C.: National Biomedical Research Foundation; 1978. pp. 345–352. volume 5. Suppl. 3 edition.
-
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. CABIOS. 1992;8:275–282. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

