GOBO: gene expression-based outcome for breast cancer online
- PMID: 21445301
- PMCID: PMC3061871
- DOI: 10.1371/journal.pone.0017911
GOBO: gene expression-based outcome for breast cancer online
Abstract
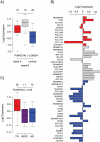
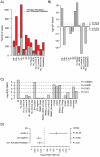
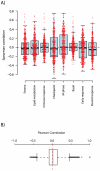
Microarray-based gene expression analysis holds promise of improving prognostication and treatment decisions for breast cancer patients. However, the heterogeneity of breast cancer emphasizes the need for validation of prognostic gene signatures in larger sample sets stratified into relevant subgroups. Here, we describe a multifunctional user-friendly online tool, GOBO (http://co.bmc.lu.se/gobo), allowing a range of different analyses to be performed in an 1881-sample breast tumor data set, and a 51-sample breast cancer cell line set, both generated on Affymetrix U133A microarrays. GOBO supports a wide range of applications including: 1) rapid assessment of gene expression levels in subgroups of breast tumors and cell lines, 2) identification of co-expressed genes for creation of potential metagenes, 3) association with outcome for gene expression levels of single genes, sets of genes, or gene signatures in multiple subgroups of the 1881-sample breast cancer data set. The design and implementation of GOBO facilitate easy incorporation of additional query functions and applications, as well as additional data sets irrespective of tumor type and array platform.
Conflict of interest statement
Figures


References
-
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
-
- Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24:4236–4244. - PubMed
-
- Farmer P, Bonnefoi H, Anderle P, Cameron D, Wirapati P, et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009;15:68–74. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

