Analysis of BAC-end sequences (BESs) and development of BES-SSR markers for genetic mapping and hybrid purity assessment in pigeonpea (Cajanus spp.)
- PMID: 21447154
- PMCID: PMC3079640
- DOI: 10.1186/1471-2229-11-56
Analysis of BAC-end sequences (BESs) and development of BES-SSR markers for genetic mapping and hybrid purity assessment in pigeonpea (Cajanus spp.)
Abstract
Background: Pigeonpea [Cajanus cajan (L.) Millsp.] is an important legume crop of rainfed agriculture. Despite of concerted research efforts directed to pigeonpea improvement, stagnated productivity of pigeonpea during last several decades may be accounted to prevalence of various biotic and abiotic constraints and the situation is exacerbated by availability of inadequate genomic resources to undertake any molecular breeding programme for accelerated crop improvement. With the objective of enhancing genomic resources for pigeonpea, this study reports for the first time, large scale development of SSR markers from BAC-end sequences and their subsequent use for genetic mapping and hybridity testing in pigeonpea.
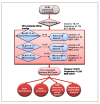
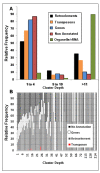
Results: A set of 88,860 BAC (bacterial artificial chromosome)-end sequences (BESs) were generated after constructing two BAC libraries by using HindIII (34,560 clones) and BamHI (34,560 clones) restriction enzymes. Clustering based on sequence identity of BESs yielded a set of >52K non-redundant sequences, comprising 35 Mbp or >4% of the pigeonpea genome. These sequences were analyzed to develop annotation lists and subdivide the BESs into genome fractions (e.g., genes, retroelements, transpons and non-annotated sequences). Parallel analysis of BESs for microsatellites or simple sequence repeats (SSRs) identified 18,149 SSRs, from which a set of 6,212 SSRs were selected for further analysis. A total of 3,072 novel SSR primer pairs were synthesized and tested for length polymorphism on a set of 22 parental genotypes of 13 mapping populations segregating for traits of interest. In total, we identified 842 polymorphic SSR markers that will have utility in pigeonpea improvement. Based on these markers, the first SSR-based genetic map comprising of 239 loci was developed for this previously uncharacterized genome. Utility of developed SSR markers was also demonstrated by identifying a set of 42 markers each for two hybrids (ICPH 2671 and ICPH 2438) for genetic purity assessment in commercial hybrid breeding programme.
Conclusion: In summary, while BAC libraries and BESs should be useful for genomics studies, BES-SSR markers, and the genetic map should be very useful for linking the genetic map with a future physical map as well as for molecular breeding in pigeonpea.
© 2011 Bohra et al; licensee BioMed Central Ltd.
Figures



References
-
- Greilhuber J, Obermayer R. Genome size variation in Cajanus cajan (Fabaceae): a reconsideration. Plant Syst Evol. 1998;212:135–141. doi: 10.1007/BF00985225. - DOI
-
- van der Maesen LJG. In: Pigeonpea. Nene YL, Hall SD, Sheila VK, editor. Wallingford: CAB International; 1990. Pigeonpea: origin, history, evolution and taxonomy; pp. 15–46.
-
- Reddy LJ, Faris DG. A cytoplasmic male sterile line in pigeonpea. International Pigeonpea Newslett. 1981;1:16–17.
-
- Marley PS, Hillocks RJ. Effect of root-knot nematodes (Meloidogyne spp.) on Fusarium wilt in pigeonpea (Cajanus cajan) Field Crop Res. 1996;46:15–20. doi: 10.1016/0378-4290(95)00083-6. - DOI
-
- Saxena KB, Sultana R, Mallikarjuna N, Saxena RK, Kumar RV, Sawargaonkar SL, Varshney RK. Male-sterility systems in pigeonpea and their role in enhancing yield. Plant Breed. 2010;129:125–134. doi: 10.1111/j.1439-0523.2009.01752.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

