UniProt Knowledgebase: a hub of integrated protein data
- PMID: 21447597
- PMCID: PMC3070428
- DOI: 10.1093/database/bar009
UniProt Knowledgebase: a hub of integrated protein data
Abstract
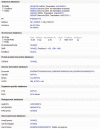
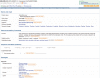
The UniProt Knowledgebase (UniProtKB) acts as a central hub of protein knowledge by providing a unified view of protein sequence and functional information. Manual and automatic annotation procedures are used to add data directly to the database while extensive cross-referencing to more than 120 external databases provides access to additional relevant information in more specialized data collections. UniProtKB also integrates a range of data from other resources. All information is attributed to its original source, allowing users to trace the provenance of all data. The UniProt Consortium is committed to using and promoting common data exchange formats and technologies, and UniProtKB data is made freely available in a range of formats to facilitate integration with other databases. Database URL: http://www.uniprot.org/
Figures



References
-
- SPIN. http://www.ebi.ac.uk/swissprot/Submissions/spin/ (17 February 2011, date last acessed)
-
- CiteXplore. http://www.ebi.ac.uk/citexplore/ (17 February 2011, date last acessed)
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

