The role of cotranscriptional histone methylations
- PMID: 21447819
- PMCID: PMC3229092
- DOI: 10.1101/sqb.2010.75.036
The role of cotranscriptional histone methylations
Abstract
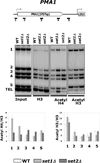
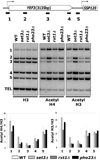
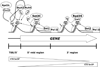
The carboxy-terminal domain (CTD) of the RNA polymerase II subunit Rpb1 undergoes dynamic phosphorylation, with different phosphorylation sites predominating at different stages of transcription. Our laboratory studies show how various mRNA-processing and chromatin-modifying enzymes interact with the phosphorylated CTD to efficiently produce mRNAs. The H3K36 methyltransferase Set2 interacts with CTD carrying phosphorylations characteristic of downstream elongation complexes, and the resulting cotranscriptional H3K36 methylation targets the Rpd3S histone deacetylase to downstream transcribed regions. Although positively correlated with gene activity, this pathway actually inhibits transcription elongation as well as initiation from cryptic promoters within genes. During early elongation, CTD serine 5 phosphorylation helps recruit the H3K4 methyltransferase complex containing Set1. Within 5' transcribed regions, cotranscriptional H3K4 dimethylation (H3K4me2) by Set1 recruits the deacetylase complex Set3C. Finally, H3K4 trimethylation at the most promoter-proximal nucleosomes is thought to stimulate transcription by promoting histone acetylation by complexes containing the ING/Yng PHD finger proteins. Surprisingly, the Rpd3L histone deacetylase complex, normally a transcription repressor, may also recognize H3K4me3. Together, the cotranscriptional histone methylations appear to function primarily to distinguish active promoter regions, which are marked by high levels of acetylation and nucleosome turnover, from the deacetylated, downstream transcribed regions of genes.
Figures
References
-
- Buratowski S. The CTD code. Nat Struct Biol. 2003;10:679–680. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
