A genomic analysis of chronological longevity factors in budding yeast
- PMID: 21447998
- PMCID: PMC3356828
- DOI: 10.4161/cc.10.9.15464
A genomic analysis of chronological longevity factors in budding yeast
Abstract
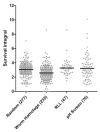
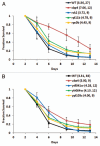
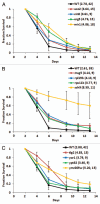
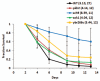
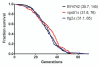
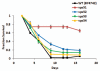
Chronological life span (CLS) has been studied as an aging paradigm in yeast. A few conserved aging genes have been identified that modulate both chronological and replicative longevity in yeast as well as longevity in the nematode Caenorhabditis elegans; however, a comprehensive analysis of the relationship between genetic control of chronological longevity and aging in other model systems has yet to be reported. To address this question, we performed a functional genomic analysis of chronological longevity for 550 single-gene deletion strains, which accounts for approximately 12% of the viable homozygous diploid deletion strains in the yeast ORF deletion collection. This study identified 33 previously unknown determinants of CLS. We found no significant enrichment for enhanced CLS among deletions corresponding to yeast orthologs of worm aging genes or among replicatively long-lived deletion strains, although a trend toward overlap was noted. In contrast, a subset of gene deletions identified from a screen for reduced acidification of culture media during growth to stationary phase was enriched for increased CLS. These results suggest that genetic control of CLS under the most commonly utilized assay conditions does not strongly overlap with longevity determinants in C. elegans, with the existing confined to a small number of genetic pathways. These data also further support the model that acidification of the culture medium plays an important role in survival during chronological aging in synthetic medium, and suggest that chronological aging studies using alternate medium conditions may be more informative with regard to aging of multicellular eukaryotes.
Figures

References
-
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91:1033–1042. - PubMed
-
- Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. - PubMed
-
- Jazwinski SM. Yeast longevity and aging—the mitochondrial connection. Mech Ageing Dev. 2005;126:243–248. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
