The deubiquitinating enzyme USP17 is essential for GTPase subcellular localization and cell motility
- PMID: 21448158
- PMCID: PMC3072070
- DOI: 10.1038/ncomms1243
The deubiquitinating enzyme USP17 is essential for GTPase subcellular localization and cell motility
Abstract
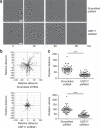
Deubiquitinating enzymes are now emerging as potential therapeutic targets that control many cellular processes, but few have been demonstrated to control cell motility. Here, we show that ubiquitin-specific protease 17 (USP17) is rapidly and transiently induced in response to chemokines SDF-1/CXCL12 and IL-8/CXCL8 in both primary cells and cell lines, and that its depletion completely blocks chemokine-induced cell migration and cytoskeletal rearrangements. Using live cell imaging, we demonstrate that USP17 is required for both elongated and amoeboid motility, in addition to chemotaxis. USP17 has previously been reported to disrupt Ras localization and we now find that USP17 depletion blocks chemokine-induced subcellular relocalization of GTPases Cdc42, Rac and RhoA, which are GTPases essential for cell motility. Collectively, these results demonstrate that USP17 has a critical role in cell migration and may be a useful drug target for both inflammatory and metastatic disease.
Figures
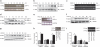





Similar articles
-
The deubiquitinating enzyme USP17 is highly expressed in tumor biopsies, is cell cycle regulated, and is required for G1-S progression.Cancer Res. 2010 Apr 15;70(8):3329-39. doi: 10.1158/0008-5472.CAN-09-4152. Epub 2010 Apr 13. Cancer Res. 2010. PMID: 20388806
-
The deubiquitinating enzyme USP17 blocks N-Ras membrane trafficking and activation but leaves K-Ras unaffected.J Biol Chem. 2010 Apr 16;285(16):12028-36. doi: 10.1074/jbc.M109.081448. Epub 2010 Feb 10. J Biol Chem. 2010. PMID: 20147298 Free PMC article.
-
A novel RCE1 isoform is required for H-Ras plasma membrane localization and is regulated by USP17.Biochem J. 2014 Jan 15;457(2):289-300. doi: 10.1042/BJ20131213. Biochem J. 2014. PMID: 24134311
-
Multifaceted role of Rho proteins in angiogenesis.J Mammary Gland Biol Neoplasia. 2005 Oct;10(4):291-8. doi: 10.1007/s10911-006-9002-8. J Mammary Gland Biol Neoplasia. 2005. PMID: 16900393 Review.
-
The Rho GTPases in macrophage motility and chemotaxis.Cell Adhes Commun. 1998;6(2-3):237-45. doi: 10.3109/15419069809004479. Cell Adhes Commun. 1998. PMID: 9823474 Review.
Cited by
-
Emerging roles of deubiquitinating enzymes in actin cytoskeleton and tumor metastasis.Cell Oncol (Dordr). 2024 Aug;47(4):1071-1089. doi: 10.1007/s13402-024-00923-z. Epub 2024 Feb 7. Cell Oncol (Dordr). 2024. PMID: 38324230 Review.
-
Deubiquitinases in cancer.Oncotarget. 2015 May 30;6(15):12872-89. doi: 10.18632/oncotarget.3671. Oncotarget. 2015. PMID: 25972356 Free PMC article. Review.
-
Evaluation of variable new antigen receptors (vNARs) as a novel cathepsin S (CTSS) targeting strategy.Front Pharmacol. 2023 Dec 5;14:1296567. doi: 10.3389/fphar.2023.1296567. eCollection 2023. Front Pharmacol. 2023. PMID: 38116078 Free PMC article.
-
USP17 Suppresses Tumorigenesis and Tumor Growth through Deubiquitinating AEP.Int J Biol Sci. 2019 Jan 29;15(4):738-748. doi: 10.7150/ijbs.30106. eCollection 2019. Int J Biol Sci. 2019. PMID: 30906206 Free PMC article.
-
The E3 deubiquitinase USP17 is a positive regulator of retinoic acid-related orphan nuclear receptor γt (RORγt) in Th17 cells.J Biol Chem. 2014 Sep 12;289(37):25546-55. doi: 10.1074/jbc.M114.565291. Epub 2014 Jul 28. J Biol Chem. 2014. PMID: 25070893 Free PMC article.
References
-
- Katz E. J., Isasa M. & Crosas B. A new map to understand deubiquitination. Biochem. Soc. Trans. 38, 21–28 (2010). - PubMed
-
- Komander D., Clague M. J. & Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 (2009). - PubMed
-
- Hussain S., Zhang Y. & Galardy P. J. DUBs and cancer: the role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle 8, 1688–1697 (2009). - PubMed
-
- Migone T. S. et al.. The deubiquitinating enzyme DUB-2 prolongs cytokine-induced signal transducers and activators of transcription activation and suppresses apoptosis following cytokine withdrawal. Blood 98, 1935–1941 (2001). - PubMed
-
- Zhu Y. et al.. DUB-2 is a member of a novel family of cytokine-inducible deubiquitinating enzymes. J. Biol. Chem. 272, 51–57 (1997). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

