The base excision repair pathway is required for efficient lentivirus integration
- PMID: 21448280
- PMCID: PMC3063173
- DOI: 10.1371/journal.pone.0017862
The base excision repair pathway is required for efficient lentivirus integration
Abstract
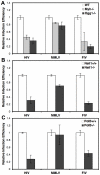
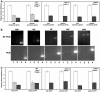
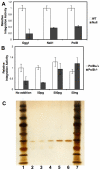
An siRNA screen has identified several proteins throughout the base excision repair (BER) pathway of oxidative DNA damage as important for efficient HIV infection. The proteins identified included early repair factors such as the base damage recognition glycosylases OGG1 and MYH and the late repair factor POLß, implicating the entire BER pathway. Murine cells with deletions of the genes Ogg1, Myh, Neil1 and Polß recapitulate the defect of HIV infection in the absence of BER. Defective infection in the absence of BER proteins was also seen with the lentivirus FIV, but not the gammaretrovirus MMLV. BER proteins do not affect HIV infection through its accessory genes nor the central polypurine tract. HIV reverse transcription and nuclear entry appear unaffected by the absence of BER proteins. However, HIV integration to the host chromosome is reduced in the absence of BER proteins. Pre-integration complexes from BER deficient cell lines show reduced integration activity in vitro. Integration activity is restored by addition of recombinant BER protein POLß. Lentiviral infection and integration efficiency appears to depend on the presence of BER proteins.
Conflict of interest statement
Figures




Similar articles
-
Repair of oxidative DNA base damage in the host genome influences the HIV integration site sequence preference.PLoS One. 2014 Jul 22;9(7):e103164. doi: 10.1371/journal.pone.0103164. eCollection 2014. PLoS One. 2014. PMID: 25051054 Free PMC article.
-
siRNA screening of a targeted library of DNA repair factors in HIV infection reveals a role for base excision repair in HIV integration.PLoS One. 2011 Mar 23;6(3):e17612. doi: 10.1371/journal.pone.0017612. PLoS One. 2011. PMID: 21448273 Free PMC article.
-
The C-terminal Domain (CTD) of Human DNA Glycosylase NEIL1 Is Required for Forming BERosome Repair Complex with DNA Replication Proteins at the Replicating Genome: DOMINANT NEGATIVE FUNCTION OF THE CTD.J Biol Chem. 2015 Aug 21;290(34):20919-20933. doi: 10.1074/jbc.M115.642918. Epub 2015 Jul 1. J Biol Chem. 2015. PMID: 26134572 Free PMC article.
-
Transcription coupled base excision repair in mammalian cells: So little is known and so much to uncover.DNA Repair (Amst). 2021 Nov;107:103204. doi: 10.1016/j.dnarep.2021.103204. Epub 2021 Aug 6. DNA Repair (Amst). 2021. PMID: 34390916 Review.
-
Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells.Cell Res. 2008 Jan;18(1):27-47. doi: 10.1038/cr.2008.8. Cell Res. 2008. PMID: 18166975 Free PMC article. Review.
Cited by
-
Both ATM and DNA-PK Are the Main Regulators of HIV-1 Post-Integrational DNA Repair.Int J Mol Sci. 2023 Feb 1;24(3):2797. doi: 10.3390/ijms24032797. Int J Mol Sci. 2023. PMID: 36769109 Free PMC article.
-
Poly(ADP-ribose) polymerase 1 promotes transcriptional repression of integrated retroviruses.J Virol. 2013 Mar;87(5):2496-507. doi: 10.1128/JVI.01668-12. Epub 2012 Dec 19. J Virol. 2013. PMID: 23255787 Free PMC article.
-
DNA strand breaks and gaps target retroviral intasome binding and integration.Nat Commun. 2023 Nov 3;14(1):7072. doi: 10.1038/s41467-023-42641-4. Nat Commun. 2023. PMID: 37923737 Free PMC article.
-
The DNA glycosylase NEIL2 is protective during SARS-CoV-2 infection.Nat Commun. 2023 Dec 9;14(1):8169. doi: 10.1038/s41467-023-43938-0. Nat Commun. 2023. PMID: 38071370 Free PMC article.
-
Host factors that promote retrotransposon integration are similar in distantly related eukaryotes.PLoS Genet. 2017 Dec 12;13(12):e1006775. doi: 10.1371/journal.pgen.1006775. eCollection 2017 Dec. PLoS Genet. 2017. PMID: 29232693 Free PMC article.
References
-
- Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1997. - PubMed
-
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. - PubMed
-
- Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

