Quantitative pharmacological analysis of antagonist binding kinetics at CRF1 receptors in vitro and in vivo
- PMID: 21449919
- PMCID: PMC3195921
- DOI: 10.1111/j.1476-5381.2011.01390.x
Quantitative pharmacological analysis of antagonist binding kinetics at CRF1 receptors in vitro and in vivo
Abstract
Background and purpose: A series of novel non-peptide corticotropin releasing factor type-1 receptor (CRF(1)) antagonists were found to display varying degrees of insurmountable and non-competitive behaviour in functional in vitro assays. We describe how we attempted to relate this behaviour to ligand receptor-binding kinetics in a quantitative manner and how this resulted in the development and implementation of an efficient pharmacological screening method based on principles described by Motulsky and Mahan.
Experimental approach: A non-equilibrium binding kinetic assay was developed to determine the receptor binding kinetics of non-peptide CRF(1) antagonists. Nonlinear, mixed-effects modelling was used to obtain estimates of the compounds association and dissociation rates. We present an integrated pharmacokinetic-pharmacodynamic (PKPD) approach, whereby the time course of in vivo CRF(1) receptor binding of novel compounds can be predicted on the basis of in vitro assays.
Key results: The non-competitive antagonist behaviour appeared to be correlated to the CRF(1) receptor off-rate kinetics. The integrated PKPD model suggested that, at least in a qualitative manner, the in vitro assay can be used to triage and select compounds for further in vivo investigations.
Conclusions and implications: This study provides evidence for a link between ligand offset kinetics and insurmountable/non-competitive antagonism at the CRF(1) receptor. The exact molecular pharmacological nature of this association remains to be determined. In addition, we have developed a quantitative framework to study and integrate in vitro and in vivo receptor binding kinetic behaviour of CRF(1) receptor antagonists in an efficient manner in a drug discovery setting.
© 2011 PFIZER Limited. British Journal of Pharmacology © 2011 The British Pharmacological Society.
Figures

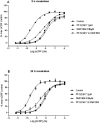
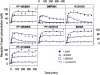


 ) and model fit (solid black line), and plasma concentration from receptor occupancy study (
) and model fit (solid black line), and plasma concentration from receptor occupancy study ( ), or satellite oral pharmacokinetic study (
), or satellite oral pharmacokinetic study ( ) and model fit (solid orange line) versus time relationships. Since DMP904 did not display a significant decay in receptor occupancy during the time course of the experiment, no attempt was made to fit a PKPD model to the data for this ligand. (B) In vivo concentration versus receptor occupancy relationship: (
) and model fit (solid orange line) versus time relationships. Since DMP904 did not display a significant decay in receptor occupancy during the time course of the experiment, no attempt was made to fit a PKPD model to the data for this ligand. (B) In vivo concentration versus receptor occupancy relationship: ( ) = observed data, solid red line = model fit. Since DMP904 did not display a significant decay in receptor occupancy during the time course of the experiment, no attempt was made to fit a PKPD model to the data for this ligand.
) = observed data, solid red line = model fit. Since DMP904 did not display a significant decay in receptor occupancy during the time course of the experiment, no attempt was made to fit a PKPD model to the data for this ligand.
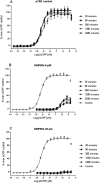
 ) and model fit (solid black line), and plasma concentration from receptor occupancy study (
) and model fit (solid black line), and plasma concentration from receptor occupancy study ( ), or satellite oral pharmacokinetic study (
), or satellite oral pharmacokinetic study ( ) and model fit (solid orange line) versus time relationships. Since DMP904 did not display a significant decay in receptor occupancy during the time course of the experiment, no attempt was made to fit a PKPD model to the data for this ligand. (B) In vivo concentration versus receptor occupancy relationship: (
) and model fit (solid orange line) versus time relationships. Since DMP904 did not display a significant decay in receptor occupancy during the time course of the experiment, no attempt was made to fit a PKPD model to the data for this ligand. (B) In vivo concentration versus receptor occupancy relationship: ( ) = observed data, solid red line = model fit. Since DMP904 did not display a significant decay in receptor occupancy during the time course of the experiment, no attempt was made to fit a PKPD model to the data for this ligand.
) = observed data, solid red line = model fit. Since DMP904 did not display a significant decay in receptor occupancy during the time course of the experiment, no attempt was made to fit a PKPD model to the data for this ligand.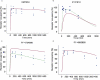
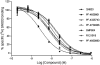
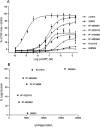
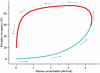
 ). (A) DMP904, (B) R121919, (C) PF-4734666, (D) PF-4850890.
). (A) DMP904, (B) R121919, (C) PF-4734666, (D) PF-4850890.
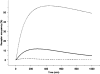
References
-
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Chalmers DT, Lovenberg TW, Grigoriadis DE, Behan DP, De Souza EB. Corticotrophin-releasing factor receptors: from molecular biology to drug design. Trends Pharmacol Sci. 1996;17:166–172. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

