Profiling metabolites and peptides in single cells
- PMID: 21451513
- PMCID: PMC3312877
- DOI: 10.1038/nmeth.1549
Profiling metabolites and peptides in single cells
Abstract
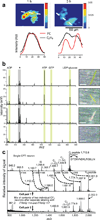
The intracellular levels and spatial localizations of metabolites and peptides reflect the state of a cell and its relationship to its surrounding environment. Moreover, the amounts and dynamics of metabolites and peptides are indicative of normal or pathological cellular conditions. Here we highlight established and evolving strategies for characterizing the metabolome and peptidome of single cells. Focused studies of the chemical composition of individual cells and functionally defined groups of cells promise to provide a greater understanding of cell fate, function and homeostatic balance. Single-cell bioanalytical microanalysis has also become increasingly valuable for examining cellular heterogeneity, particularly in the fields of neuroscience, stem cell biology and developmental biology.
Figures


References
-
- Hausser M. Nat. Neurosci. 2000;3 Suppl:1165. - PubMed
-
- Hubel David H, Wiesel Torsten N. Brain and visual perception: the story of a 25-year collaboration. New York, N.Y.: Oxford University Press; 2005.
-
- Moroz LL, Gillette R, Sweedler JV. J. Exp. Biol. 1999;202:333–341. - PubMed
-
- Brecht M, Schneider M, Sakmann B, et al. Nature. 2004;427:704–710. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

