Large-scale 13C-flux analysis reveals distinct transcriptional control of respiratory and fermentative metabolism in Escherichia coli
- PMID: 21451587
- PMCID: PMC3094070
- DOI: 10.1038/msb.2011.9
Large-scale 13C-flux analysis reveals distinct transcriptional control of respiratory and fermentative metabolism in Escherichia coli
Abstract
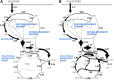
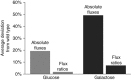
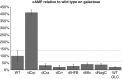
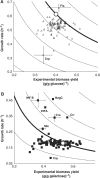
Despite our increasing topological knowledge on regulation networks in model bacteria, it is largely unknown which of the many co-occurring regulatory events actually control metabolic function and the distribution of intracellular fluxes. Here, we unravel condition-dependent transcriptional control of Escherichia coli metabolism by large-scale (13)C-flux analysis in 91 transcriptional regulator mutants on glucose and galactose. In contrast to the canonical respiro-fermentative glucose metabolism, fully respiratory galactose metabolism depends exclusively on the phosphoenol-pyruvate (PEP)-glyoxylate cycle. While 2/3 of the regulators directly or indirectly affected absolute flux rates, the partitioning between different pathways remained largely stable with transcriptional control focusing primarily on the acetyl-CoA branch point. Flux distribution control was achieved by nine transcription factors on glucose, including ArcA, Fur, PdhR, IHF A and IHF B, but was exclusively mediated by the cAMP-dependent Crp regulation of the PEP-glyoxylate cycle flux on galactose. Five further transcription factors affected this flux only indirectly through cAMP and Crp by increasing the galactose uptake rate. Thus, E. coli actively limits its galactose catabolism at the expense of otherwise possible faster growth.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Comment in
-
Transcriptional control of metabolic fluxes.Mol Syst Biol. 2011 Mar 29;7:478. doi: 10.1038/msb.2011.10. Mol Syst Biol. 2011. PMID: 21451588 Free PMC article. No abstract available.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

