Immunological network signatures of cancer progression and survival
- PMID: 21453479
- PMCID: PMC3094196
- DOI: 10.1186/1755-8794-4-28
Immunological network signatures of cancer progression and survival
Abstract
Background: The immune contribution to cancer progression is complex and difficult to characterize. For example in tumors, immune gene expression is detected from the combination of normal, tumor and immune cells in the tumor microenvironment. Profiling the immune component of tumors may facilitate the characterization of the poorly understood roles immunity plays in cancer progression. However, the current approaches to analyze the immune component of a tumor rely on incomplete identification of immune factors.
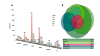
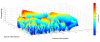
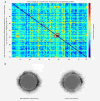
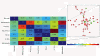
Methods: To facilitate a more comprehensive approach, we created a ranked immunological relevance score for all human genes, developed using a novel strategy that combines text mining and information theory. We used this score to assign an immunological grade to gene expression profiles, and thereby quantify the immunological component of tumors. This immunological relevance score was benchmarked against existing manually curated immune resources as well as high-throughput studies. To further characterize immunological relevance for genes, the relevance score was charted against both the human interactome and cancer information, forming an expanded interactome landscape of tumor immunity. We applied this approach to expression profiles in melanomas, thus identifying and grading their immunological components, followed by identification of their associated protein interactions.
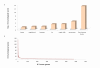
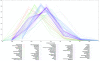
Results: The power of this strategy was demonstrated by the observation of early activation of the adaptive immune response and the diversity of the immune component during melanoma progression. Furthermore, the genome-wide immunological relevance score classified melanoma patient groups, whose immunological grade correlated with clinical features, such as immune phenotypes and survival.
Conclusions: The assignment of a ranked immunological relevance score to all human genes extends the content of existing immune gene resources and enriches our understanding of immune involvement in complex biological networks. The application of this approach to tumor immunity represents an automated systems strategy that quantifies the immunological component in complex disease. In so doing, it stratifies patients according to their immune profiles, which may lead to effective computational prognostic and clinical guides.
Figures
References
-
- Virchow RL. Die Krankhaften Geschwülste Dreissig Vorlesungen gehalten während des Wintersemesters 1862-63 Vierte Vorlesung. Vol. 1. Berlin; 1863. p. 65.
-
- Bogunovic D, O'Neill DW, Belitskaya-Levy I, Vacic V, Yu YL, Adams S, Darvishian F, Berman R, Shapiro R, Pavlick AC. et al.Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc Natl Acad Sci USA. 2009;106:20429–20434. doi: 10.1073/pnas.0905139106. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

