The loop-less tmCdc34 E2 mutant defective in polyubiquitination in vitro and in vivo supports yeast growth in a manner dependent on Ubp14 and Cka2
- PMID: 21453497
- PMCID: PMC3080790
- DOI: 10.1186/1747-1028-6-7
The loop-less tmCdc34 E2 mutant defective in polyubiquitination in vitro and in vivo supports yeast growth in a manner dependent on Ubp14 and Cka2
Erratum in
-
Erratum to: The loop-less tmCdc34 E2 mutant defective polyubiquitination in vitro and in vivo supports yeast growth in a manner dependent on Ubp14 and Cka2.Cell Div. 2016 Oct 6;11:13. doi: 10.1186/s13008-016-0018-1. eCollection 2016. Cell Div. 2016. PMID: 27761151 Free PMC article.
Abstract
Background: The S73/S97/loop motif is a hallmark of the Cdc34 family of E2 ubiquitin-conjugating enzymes that together with the SCF E3 ubiquitin ligases promote degradation of proteins involved in cell cycle and growth regulation. The inability of the loop-less Δ12Cdc34 mutant to support growth was linked to its inability to catalyze polyubiquitination. However, the loop-less triple mutant (tm) Cdc34, which not only lacks the loop but also contains the S73K and S97D substitutions typical of the K73/D97/no loop motif present in other E2s, supports growth. Whether tmCdc34 supports growth despite defective polyubiquitination, or the S73K and S97D substitutions, directly or indirectly, correct the defect caused by the loop absence, are unknown.
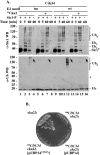
Results: tmCdc34 supports yeast viability with normal cell size and cell cycle profile despite producing fewer polyubiquitin conjugates in vivo and in vitro. The in vitro defect in Sic1 substrate polyubiquitination is similar to the defect observed in reactions with Δ12Cdc34 that cannot support growth. The synthesis of free polyubiquitin by tmCdc34 is activated only modestly and in a manner dependent on substrate recruitment to SCFCdc4. Phosphorylation of C-terminal serines in tmCdc34 by Cka2 kinase prevents the synthesis of free polyubiquitin chains, likely by promoting their attachment to substrate. Nevertheless, tmCDC34 yeast are sensitive to loss of the Ubp14 C-terminal ubiquitin hydrolase and DUBs other than Ubp14 inefficiently disassemble polyubiquitin chains produced in tmCDC34 yeast extracts, suggesting that the free chains, either synthesized de novo or recycled from substrates, have an altered structure.
Conclusions: The catalytic motif replacement compromises polyubiquitination activity of Cdc34 and alters its regulation in vitro and in vivo, but either motif can support Cdc34 function in yeast viability. Robust polyubiquitination mediated by the S73/S97/loop motif is thus not necessary for Cdc34 role in yeast viability, at least under typical laboratory conditions.
Figures

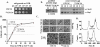




References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

