Using context to improve protein domain identification
- PMID: 21453511
- PMCID: PMC3090354
- DOI: 10.1186/1471-2105-12-90
Using context to improve protein domain identification
Abstract
Background: Identifying domains in protein sequences is an important step in protein structural and functional annotation. Existing domain recognition methods typically evaluate each domain prediction independently of the rest. However, the majority of proteins are multidomain, and pairwise domain co-occurrences are highly specific and non-transitive.
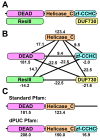
Results: Here, we demonstrate how to exploit domain co-occurrence to boost weak domain predictions that appear in previously observed combinations, while penalizing higher confidence domains if such combinations have never been observed. Our framework, Domain Prediction Using Context (dPUC), incorporates pairwise "context" scores between domains, along with traditional domain scores and thresholds, and improves domain prediction across a variety of organisms from bacteria to protozoa and metazoa. Among the genomes we tested, dPUC is most successful at improving predictions for the poorly-annotated malaria parasite Plasmodium falciparum, for which over 38% of the genome is currently unannotated. Our approach enables high-confidence annotations in this organism and the identification of orthologs to many core machinery proteins conserved in all eukaryotes, including those involved in ribosomal assembly and other RNA processing events, which surprisingly had not been previously known.
Conclusions: Overall, our results demonstrate that this new context-based approach will provide significant improvements in domain and function prediction, especially for poorly understood genomes for which the need for additional annotations is greatest. Source code for the algorithm is available under a GPL open source license at http://compbio.cs.princeton.edu/dpuc/. Pre-computed results for our test organisms and a web server are also available at that location.
Figures

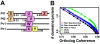
References
-
- Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, Bryant SH. CDD: specific functional annotation with the Conserved Domain Database. Nucl Acids Res. 2009;37:D205–210. doi: 10.1093/nar/gkn845. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

