Derivatives of rhodamine 19 as mild mitochondria-targeted cationic uncouplers
- PMID: 21454507
- PMCID: PMC3093858
- DOI: 10.1074/jbc.M110.212837
Derivatives of rhodamine 19 as mild mitochondria-targeted cationic uncouplers
Abstract
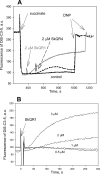
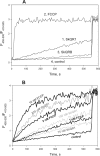
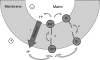
A limited decrease in mitochondrial membrane potential can be beneficial for cells, especially under some pathological conditions, suggesting that mild uncouplers (protonophores) causing such an effect are promising candidates for therapeutic uses. The great majority of protonophores are weak acids capable of permeating across membranes in their neutral and anionic forms. In the present study, protonophorous activity of a series of derivatives of cationic rhodamine 19, including dodecylrhodamine (C(12)R1) and its conjugate with plastoquinone (SkQR1), was revealed using a variety of assays. Derivatives of rhodamine B, lacking dissociable protons, showed no protonophorous properties. In planar bilayer lipid membranes, separating two compartments differing in pH, diffusion potential of H(+) ions was generated in the presence of C(12)R1 and SkQR1. These compounds induced pH equilibration in liposomes loaded with the pH probe pyranine. C(12)R1 and SkQR1 partially stimulated respiration of rat liver mitochondria in State 4 and decreased their membrane potential. Also, C(12)R1 partially stimulated respiration of yeast cells but, unlike the anionic protonophore FCCP, did not suppress their growth. Loss of function of mitochondrial DNA in yeast (grande-petite transformation) is known to cause a major decrease in the mitochondrial membrane potential. We found that petite yeast cells are relatively more sensitive to the anionic uncouplers than to C(12)R1 compared with grande cells. Together, our data suggest that rhodamine 19-based cationic protonophores are self-limiting; their uncoupling activity is maximal at high membrane potential, but the activity decreases membrane potentials, which causes partial efflux of the uncouplers from mitochondria and, hence, prevents further membrane potential decrease.
© 2011 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






Similar articles
-
Penetrating cations enhance uncoupling activity of anionic protonophores in mitochondria.PLoS One. 2013 Apr 23;8(4):e61902. doi: 10.1371/journal.pone.0061902. Print 2013. PLoS One. 2013. PMID: 23626747 Free PMC article.
-
Electrogenic proton transport across lipid bilayer membranes mediated by cationic derivatives of rhodamine 19: comparison with anionic protonophores.Eur Biophys J. 2013 Jun;42(6):477-85. doi: 10.1007/s00249-013-0898-9. Epub 2013 Apr 5. Eur Biophys J. 2013. PMID: 23558512
-
Weak C-H acids as protonophores can carry hydrogen ions through lipid membranes and mitochondria: a case of o-carborane.Phys Chem Chem Phys. 2016 Jun 28;18(24):16476-82. doi: 10.1039/c6cp02581a. Epub 2016 Jun 6. Phys Chem Chem Phys. 2016. PMID: 27265316
-
Novel penetrating cations for targeting mitochondria.Curr Pharm Des. 2013;19(15):2795-806. doi: 10.2174/1381612811319150015. Curr Pharm Des. 2013. PMID: 23092317 Review.
-
Uncouplers of oxidative phosphorylation.Environ Health Perspect. 1990 Jul;87:213-8. doi: 10.1289/ehp.9087213. Environ Health Perspect. 1990. PMID: 2176586 Free PMC article. Review.
Cited by
-
Mitochondria-targeted triphenylphosphonium-based compounds do not affect estrogen receptor α.PeerJ. 2020 Mar 25;8:e8803. doi: 10.7717/peerj.8803. eCollection 2020. PeerJ. 2020. PMID: 32257641 Free PMC article.
-
Ionophoric effects of the antitubercular drug bedaquiline.Proc Natl Acad Sci U S A. 2018 Jul 10;115(28):7326-7331. doi: 10.1073/pnas.1803723115. Epub 2018 Jun 25. Proc Natl Acad Sci U S A. 2018. PMID: 29941569 Free PMC article.
-
The impact of mitochondrial dysfunction on osteoarthritis cartilage: current insights and emerging mitochondria-targeted therapies.Bone Res. 2025 Sep 1;13(1):77. doi: 10.1038/s41413-025-00460-x. Bone Res. 2025. PMID: 40887466 Free PMC article. Review.
-
Penetrating cations induce pleiotropic drug resistance in yeast.Sci Rep. 2018 May 25;8(1):8131. doi: 10.1038/s41598-018-26435-z. Sci Rep. 2018. PMID: 29802261 Free PMC article.
-
Effect of liposomes on energy-dependent uptake of the antioxidant SkQR1 by isolated mitochondria.J Bioenerg Biomembr. 2012 Aug;44(4):453-60. doi: 10.1007/s10863-012-9449-9. Epub 2012 Jun 22. J Bioenerg Biomembr. 2012. PMID: 22723179
References
-
- Korshunov S. S., Skulachev V. P., Starkov A. A. (1997) FEBS Lett. 416, 15–18 - PubMed
-
- Skulachev V. P. (1996) Q. Rev. Biophys. 29, 169–202 - PubMed
-
- Nedergaard J., Cannon B. (2003) Exp. Physiol. 88, 65–84 - PubMed
-
- Blaikie F. H., Brown S. E., Samuelsson L. M., Brand M. D., Smith R. A., Murphy M. P. (2006) Biosci. Rep. 26, 231–243 - PubMed
-
- Harper J. A., Dickinson K., Brand M. D. (2001) Obes. Rev. 2, 255–265 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

