Retinyl ester storage particles (retinosomes) from the retinal pigmented epithelium resemble lipid droplets in other tissues
- PMID: 21454509
- PMCID: PMC3089567
- DOI: 10.1074/jbc.M110.195198
Retinyl ester storage particles (retinosomes) from the retinal pigmented epithelium resemble lipid droplets in other tissues
Abstract
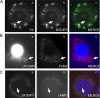
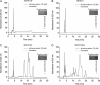
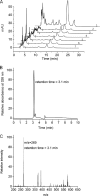
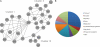
Levels of many hydrophobic cellular substances are tightly regulated because of their potential cytotoxicity. These compounds tend to self-aggregate in cytoplasmic storage depots termed lipid droplets/bodies that have well defined structures that contain additional components, including cholesterol and various proteins. Hydrophobic substances in these structures become mobilized in a specific and regulated manner as dictated by cellular requirements. Retinal pigmented epithelial cells in the eye produce retinyl ester-containing lipid droplets named retinosomes. These esters are mobilized to replenish the visual chromophore, 11-cis-retinal, and their storage ensures proper visual function despite fluctuations in dietary vitamin A intake. But it remains unclear whether retinosomes are structures specific to the eye or similar to lipid droplets in other organs/tissues that contain substances other than retinyl esters. Thus, we initially investigated the production of these lipid droplets in experimental cell lines expressing lecithin:retinol acyltransferase, a key enzyme involved in formation of retinyl ester-containing retinosomes from all-trans-retinol. We found that retinosomes and oleate-derived lipid droplets form and co-localize concomitantly, indicating their intrinsic structural similarities. Next, we isolated native retinosomes from bovine retinal pigmented epithelium and found that their protein and hydrophobic small molecular constituents were similar to those of lipid droplets reported for other experimental cell lines and tissues. These unexpected findings suggest a common mechanism for lipid droplet formation that exhibits broad chemical specificity for the hydrophobic substances being stored.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

