The human adenocarcinoma-associated gene, AGR2, induces expression of amphiregulin through Hippo pathway co-activator YAP1 activation
- PMID: 21454516
- PMCID: PMC3093902
- DOI: 10.1074/jbc.M110.215707
The human adenocarcinoma-associated gene, AGR2, induces expression of amphiregulin through Hippo pathway co-activator YAP1 activation
Abstract
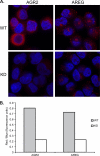
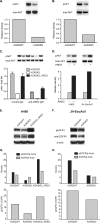
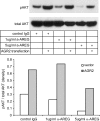
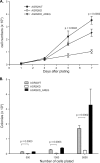
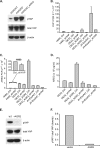
Anterior Gradient Homolog 2 (AGR2) is expressed by the normal intestine and by most human adenocarcinomas, including those derived from the esophagus, pancreas, lung, breast, ovary, and prostate. Xenografts of human adenocarcinoma cell lines in nude mice previously demonstrated that AGR2 supports tumor growth. In addition, AGR2 is able to induce in vitro a transformed phenotype in fibroblast and epithelial cell lines. The mechanism underlying the growth promoting effects of AGR2 is unknown. The present study shows that AGR2 induces expression of amphiregulin (AREG), a growth promoting EGFR ligand. Induced AREG expression in adenocarcinoma cells is able to rescue the transformed phenotype that is lost when AGR2 expression is reduced. Additional experiments demonstrate that AGR2 induction of AREG is mediated by activation of the Hippo signaling pathway co-activator, YAP1. Thus AGR2 promotes growth by regulating the Hippo and EGF receptor signaling pathways.
© 2011 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


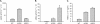


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

