Sequential establishment of marks on soluble histones H3 and H4
- PMID: 21454524
- PMCID: PMC3093847
- DOI: 10.1074/jbc.M111.223453
Sequential establishment of marks on soluble histones H3 and H4
Abstract
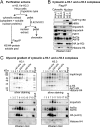
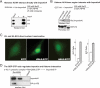
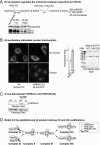
Much progress has been made concerning histone function in the nucleus; however, following their synthesis, how their marking and subcellular trafficking are regulated remains to be explored. To gain an insight into these issues, we focused on soluble histones and analyzed endogenous and tagged H3 histones in parallel. We distinguished six complexes that we could place to account for maturation events occurring on histones H3 and H4 from their synthesis onward. In each complex, a different set of chaperones is involved, and we found specific post-translational modifications. Interestingly, we revealed that histones H3 and H4 are transiently poly(ADP-ribosylated). The impact of these marks in histone metabolism proved to be important as we found that acetylation of lysines 5 and 12 on histone H4 stimulated its nuclear translocation. Furthermore, we showed that, depending on particular histone H3 modifications, the balance in the presence of the different translocation complexes changes. Therefore, our results enabled us to propose a regulatory means of these marks for controlling cytoplasmic/nuclear shuttling and the establishment of early modification patterns.
© 2011 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

