Sequence and generation of mature ribosomal RNA transcripts in Dictyostelium discoideum
- PMID: 21454536
- PMCID: PMC3093845
- DOI: 10.1074/jbc.M110.208306
Sequence and generation of mature ribosomal RNA transcripts in Dictyostelium discoideum
Abstract
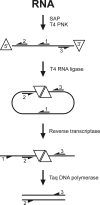
The amoeba Dictyostelium discoideum is a well established model organism for studying numerous aspects of cellular and developmental functions. Its ribosomal RNA (rRNA) is encoded in an extrachromosomal palindrome that exists in ∼100 copies in the cell. In this study, we have set out to investigate the sequence of the expressed rRNA. For this, we have ligated the rRNA ends and performed RT-PCR on these circular RNAs. Sequencing revealed that the mature 26 S, 17 S, 5.8 S, and 5 S rRNAs have sizes of 3741, 1871, 162, and 112 nucleotides, respectively. Unlike the published data, all mature rRNAs of the same type uniformly display the same start and end nucleotides in the analyzed AX2 strain. We show the existence of a short lived primary transcript covering the rRNA transcription unit of 17 S, 5.8 S, and 26 S rRNA. Northern blots and RT-PCR reveal that from this primary transcript two precursor molecules of the 17 S and two precursors of the 26 S rRNA are generated. We have also determined the sequences of these precursor molecules, and based on these data, we propose a model for the maturation of the rRNAs in Dictyostelium discoideum that we compare with the processing of the rRNA transcription unit of Saccharomyces cerevisiae.
© 2011 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

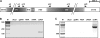




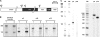


References
-
- Nissen P., Hansen J., Ban N., Moore P. B., Steitz T. A. (2000) Science 289, 920–930 - PubMed
-
- Schluenzen F., Tocilj A., Zarivach R., Harms J., Gluehmann M., Janell D., Bashan A., Bartels H., Agmon I., Franceschi F., Yonath A. (2000) Cell 102, 615–623 - PubMed
-
- Wimberly B. T., Brodersen D. E., Clemons W. M., Jr., Morgan-Warren R. J., Carter A. P., Vonrhein C., Hartsch T., Ramakrishnan V. (2000) Nature 407, 327–339 - PubMed
-
- Noller H. F., Hoffarth V., Zimniak L. (1992) Science 256, 1416–1419 - PubMed
-
- Maizels N. (1976) Cell 9, 431–438 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

