Regulatory circuits of the AAA+ disaggregase Hsp104
- PMID: 21454552
- PMCID: PMC3093873
- DOI: 10.1074/jbc.M110.216176
Regulatory circuits of the AAA+ disaggregase Hsp104
Abstract
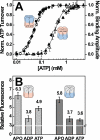
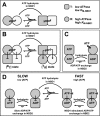
Yeast Hsp104 is an AAA+ chaperone that rescues proteins from the aggregated state. Six protomers associate to form the functional hexamer. Each protomer contains two AAA+ modules, NBD1 and NBD2. Hsp104 converts energy provided by ATP into mechanical force used to thread polypeptides through its axial channel, thereby disrupting protein aggregates. But how the action of its 12 AAA+ domains is co-ordinated to catalyze disaggregation remained unexplained. Here, we identify a sophisticated allosteric network consisting of three distinct pathways that senses the nucleotide state of AAA+ modules and transmits this information across the Hsp104 hexamer. As a result of this communication, NBD1 and NBD2 each adopt two distinct conformations (relaxed and tense) that are reciprocally regulated. The key element in the network is the NBD1-ATP state that enables Hsp104 to switch from a barely active [(T)(R)] state to a highly active [(R)(T)] state. This concerted switch involves both cis and trans protomer interactions and provides Hsp104 with the mechanistic scaffold to catalyze disaggregation. It prepares the chaperone for polypeptide binding and activates NBD2 to generate the power strokes required to resolve protein aggregates. ATP hydrolysis in NBD1 resolves the high affinity [(R)(T)] state and switches the chaperone back into the low affinity [(T)(R)] state. Our model integrates previously unexplained observations and provides the first comprehensive map of nucleotide-related allosteric signals in a class-1 AAA+ protein.
© 2011 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Ogura T., Wilkinson A. J. (2001) Genes Cells 6, 575–597 - PubMed
-
- Erzberger J. P., Berger J. M. (2006) Annu. Rev. Biophys. Biomol. Struct. 35, 93–114 - PubMed
-
- Parsell D. A., Kowal A. S., Singer M. A., Lindquist S. (1994) Nature 372, 475–478 - PubMed
-
- Sanchez Y., Lindquist S. L. (1990) Science 248, 1112–1115 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

