A genetic screen targeted on the FO component of mitochondrial ATP synthase in Saccharomyces cerevisiae
- PMID: 21454598
- PMCID: PMC3093890
- DOI: 10.1074/jbc.M110.214825
A genetic screen targeted on the FO component of mitochondrial ATP synthase in Saccharomyces cerevisiae
Abstract
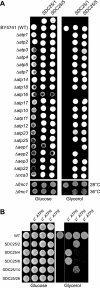
In yeast, the two main F(O) proton-translocating subunits of the ATP synthase (subunits 6/a and 9/c) are encoded by mitochondrial DNA (mtDNA). Unfortunately, mutations that inactivate the F(O) typically result in loss of mtDNA under the form of ρ(-)/ρ(0) cells. Thus, we have designed a novel genetic strategy to circumvent this problem. It exploits previous findings that a null mutation in the nuclear ATP16 gene encoding ATP synthase subunit δ results in massive and lethal F(O)-mediated protons leaks across the inner mitochondrial membrane. Mutations that inactivate the F(O) can thus, in these conditions, be selected positively as cell viability rescuing events. A first set of seven mutants was analyzed and all showed, as expected, very severe F(O) deficiencies. Two mutants carried nuclear mutations in known genes (AEP1, AEP2) required for subunit c expression. The five other mutations were located in mtDNA. Of these, three affect synthesis or stability of subunit a transcripts and the two last consisted in a single amino acid replacement in subunit c. One of the subunit c mutations is particularly interesting. It consists in an alanine to valine change at position 60 of subunit c adjacent to the essential glutamate of subunit c (at position 59) that interacts with the essential arginine 186 of subunit a. The properties of this mutant suggest that the contact zone between subunit a and the ten subunits c-ring structure only involves critical transient interactions confined to the region where protons are exchanged between the subunit a and the c-ring.
© 2011 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Boyer P. D. (1997) Annu. Rev. Biochem. 66, 717–749 - PubMed
-
- Ackerman S. H., Tzagoloff A. (2005) Prog. Nucleic Acids Res. Mol. Biol. 80, 95–133 - PubMed
-
- Senior A. E., Nadanaciva S., Weber J. (2002) Biochim. Biophys. Acta 1553, 188–211 - PubMed
-
- Abrahams J. P., Leslie A. G., Lutter R., Walker J. E. (1994) Nature 370, 621–628 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

