Three-dimensional root phenotyping with a novel imaging and software platform
- PMID: 21454799
- PMCID: PMC3177249
- DOI: 10.1104/pp.110.169102
Three-dimensional root phenotyping with a novel imaging and software platform
Abstract
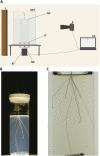
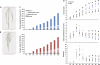
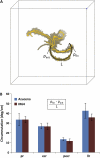
A novel imaging and software platform was developed for the high-throughput phenotyping of three-dimensional root traits during seedling development. To demonstrate the platform's capacity, plants of two rice (Oryza sativa) genotypes, Azucena and IR64, were grown in a transparent gellan gum system and imaged daily for 10 d. Rotational image sequences consisting of 40 two-dimensional images were captured using an optically corrected digital imaging system. Three-dimensional root reconstructions were generated and analyzed using a custom-designed software, RootReader3D. Using the automated and interactive capabilities of RootReader3D, five rice root types were classified and 27 phenotypic root traits were measured to characterize these two genotypes. Where possible, measurements from the three-dimensional platform were validated and were highly correlated with conventional two-dimensional measurements. When comparing gellan gum-grown plants with those grown under hydroponic and sand culture, significant differences were detected in morphological root traits (P < 0.05). This highly flexible platform provides the capacity to measure root traits with a high degree of spatial and temporal resolution and will facilitate novel investigations into the development of entire root systems or selected components of root systems. In combination with the extensive genetic resources that are now available, this platform will be a powerful resource to further explore the molecular and genetic determinants of root system architecture.
Figures


References
-
- Armengaud P, Zambaux K, Hills A, Sulpice R, Pattison RJ, Blatt MR, Amtmann A. (2009) EZ-Rhizo: integrated software for the fast and accurate measurement of root system architecture. Plant J 57: 945–956 - PubMed
-
- Berntson GM. (1994) Modelling root architecture: are there tradeoffs between efficiency and potential of resource acquisition? New Phytol 127: 483–493
-
- de Dorlodot S, Forster B, Pagès L, Price A, Tuberosa R, Draye X. (2007) Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci 12: 474–481 - PubMed
-
- Eissenstat DM. (1991) On the relationship between specific root length and rate of root proliferation: a field study using citrus rootstocks. New Phytol 118: 63–68
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

