Phenobarbital mediates an epigenetic switch at the constitutive androstane receptor (CAR) target gene Cyp2b10 in the liver of B6C3F1 mice
- PMID: 21455306
- PMCID: PMC3063791
- DOI: 10.1371/journal.pone.0018216
Phenobarbital mediates an epigenetic switch at the constitutive androstane receptor (CAR) target gene Cyp2b10 in the liver of B6C3F1 mice
Abstract
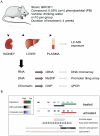
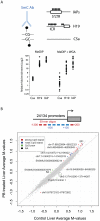
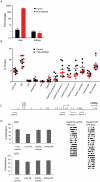
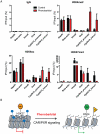
Evidence suggests that epigenetic perturbations are involved in the adverse effects associated with some drugs and toxicants, including certain classes of non-genotoxic carcinogens. Such epigenetic changes (altered DNA methylation and covalent histone modifications) may take place at the earliest stages of carcinogenesis and their identification holds great promise for biomedical research. Here, we evaluate the sensitivity and specificity of genome-wide epigenomic and transcriptomic profiling in phenobarbital (PB)-treated B6C3F1 mice, a well-characterized rodent model of non-genotoxic liver carcinogenesis. Methylated DNA Immunoprecipitation (MeDIP)-coupled microarray profiling of 17,967 promoter regions and 4,566 intergenic CpG islands was combined with genome-wide mRNA expression profiling to identify liver tissue-specific PB-mediated DNA methylation and transcriptional alterations. Only a limited number of significant anti-correlations were observed between PB-induced transcriptional and promoter-based DNA methylation perturbations. However, the constitutive androstane receptor (CAR) target gene Cyp2b10 was found to be concomitantly hypomethylated and transcriptionally activated in a liver tissue-specific manner following PB treatment. Furthermore, analysis of active and repressive histone modifications using chromatin immunoprecipitation revealed a strong PB-mediated epigenetic switch at the Cyp2b10 promoter. Our data reveal that PB-induced transcriptional perturbations are not generally associated with broad changes in the DNA methylation status at proximal promoters and suggest that the drug-inducible CAR pathway regulates an epigenetic switch from repressive to active chromatin at the target gene Cyp2b10. This study demonstrates the utility of integrated epigenomic and transcriptomic profiling for elucidating early mechanisms and biomarkers of non-genotoxic carcinogenesis.
Conflict of interest statement
Figures
Similar articles
-
The roles of co-chaperone CCRP/DNAJC7 in Cyp2b10 gene activation and steatosis development in mouse livers.PLoS One. 2014 Dec 26;9(12):e115663. doi: 10.1371/journal.pone.0115663. eCollection 2014. PLoS One. 2014. PMID: 25542016 Free PMC article.
-
Protein Kinase N Family Negatively Regulates Constitutive Androstane Receptor-Mediated Transcriptional Induction of Cytochrome P450 2b10 in the Livers of Mice.J Pharmacol Exp Ther. 2021 Oct;379(1):53-63. doi: 10.1124/jpet.121.000790. Epub 2021 Jul 26. J Pharmacol Exp Ther. 2021. PMID: 34312179
-
Constitutive androstane receptor (CAR) mediates dieldrin-induced liver tumorigenesis in mouse.Arch Toxicol. 2020 Aug;94(8):2873-2884. doi: 10.1007/s00204-020-02781-8. Epub 2020 May 20. Arch Toxicol. 2020. PMID: 32435917
-
Mode of action and human relevance analysis for nuclear receptor-mediated liver toxicity: A case study with phenobarbital as a model constitutive androstane receptor (CAR) activator.Crit Rev Toxicol. 2014 Jan;44(1):64-82. doi: 10.3109/10408444.2013.835786. Epub 2013 Nov 4. Crit Rev Toxicol. 2014. PMID: 24180433 Free PMC article. Review.
-
Phenobarbital induction of drug/steroid-metabolizing enzymes and nuclear receptor CAR.Biochim Biophys Acta. 2003 Feb 17;1619(3):239-42. doi: 10.1016/s0304-4165(02)00482-8. Biochim Biophys Acta. 2003. PMID: 12573483 Review.
Cited by
-
Impact of Neonatal Activation of Nuclear Receptor CAR (Nr1i3) on Cyp2 Gene Expression in Adult Mouse Liver.Toxicol Sci. 2022 May 26;187(2):298-310. doi: 10.1093/toxsci/kfac032. Toxicol Sci. 2022. PMID: 35285501 Free PMC article.
-
Gene and genome parameters of mammalian liver circadian genes (LCGs).PLoS One. 2012;7(10):e46961. doi: 10.1371/journal.pone.0046961. Epub 2012 Oct 10. PLoS One. 2012. PMID: 23071677 Free PMC article.
-
Emphasising the European Union's Commitment to Cancer Research: a helicopter view of the Seventh Framework Programme for Research and Technological Development.Oncologist. 2012;17(10):e26-32. doi: 10.1634/theoncologist.2012-0327. Oncologist. 2012. PMID: 23104172 Free PMC article.
-
Incorporation of an Epigenetic Evaluation into Safety Assessment: What we First Need to Know.Curr Opin Toxicol. 2017 Apr;3:20-24. doi: 10.1016/j.cotox.2017.05.001. Epub 2017 May 5. Curr Opin Toxicol. 2017. PMID: 30740577 Free PMC article.
-
Epigenetic Mechanisms Contribute to Intraindividual Variations of Drug Metabolism Mediated by Cytochrome P450 Enzymes.Drug Metab Dispos. 2023 Jun;51(6):672-684. doi: 10.1124/dmd.122.001007. Epub 2023 Mar 27. Drug Metab Dispos. 2023. PMID: 36973001 Free PMC article. Review.
References
-
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. - PubMed
-
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. - PubMed
-
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. - PubMed
-
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. - PubMed
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

