Reproducing crystal binding modes of ligand functional groups using Site-Identification by Ligand Competitive Saturation (SILCS) simulations
- PMID: 21456594
- PMCID: PMC3090225
- DOI: 10.1021/ci100462t
Reproducing crystal binding modes of ligand functional groups using Site-Identification by Ligand Competitive Saturation (SILCS) simulations
Abstract
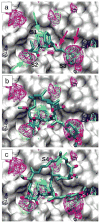
The applicability of a computational method, Site Identification by Ligand Competitive Saturation (SILCS), to identify regions on a protein surface with which different types of functional groups on low-molecular weight inhibitors interact is demonstrated. The method involves molecular dynamics (MD) simulations of a protein in an aqueous solution of chemically diverse small molecules from which probability distributions of fragments types, termed FragMaps, are obtained. In the present application, SILCS simulations are performed with an aqueous solution of 1 M benzene and propane to map the affinity pattern of the protein for aromatic and aliphatic functional groups. In addition, water hydrogen and oxygen atoms serve as probes for hydrogen-bond donor and acceptor affinity, respectively. The method is tested using a set of 7 proteins for which crystal structures of complexes with several high affinity inhibitors are known. Good agreement is obtained between FragMaps and the positions of chemically similar functional groups in inhibitors as observed in the X-ray crystallographic structures. Quantitative capabilities of the SILCS approach are demonstrated by converting FragMaps to free energies, termed Grid Free Energies (GFE), and showing correlation between the GFE values and experimental binding affinities. For proteins for which ligand decoy sets are available, GFE values are shown to typically score the crystal conformation and conformations similar to it more favorable than decoys. Additionally, SILCS is tested for its ability to capture the subtle differences in ligand affinity across homologous proteins, information which may be of utility toward specificity-guided drug design. Taken together, our results show that SILCS can recapitulate the known location of functional groups of bound inhibitors for a number of proteins, suggesting that the method may be of utility for rational drug design.
Figures










References
-
- Jorgensen WL. The many roles of computation in drug discovery. Science. 2004;303:1813–1818. - PubMed
-
- Congreve M, Chessari G, Tisi D, Woodhead AJ. Recent developments in fragment-based drug discovery. J Med Chem. 2008;51:3661–3680. - PubMed
-
- Kollman PA. Free energy calculations: Applications to chemical and biochemical phenomena. Chem Rev. 1993;93:2395–2417.
-
- Erlanson DA, McDowell RS, O’Brien T. Fragment-based drug discovery. J Med Chem. 2004;47:3463–3482. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

