Identification and characterisation of seed storage protein transcripts from Lupinus angustifolius
- PMID: 21457583
- PMCID: PMC3078879
- DOI: 10.1186/1471-2229-11-59
Identification and characterisation of seed storage protein transcripts from Lupinus angustifolius
Abstract
Background: In legumes, seed storage proteins are important for the developing seedling and are an important source of protein for humans and animals. Lupinus angustifolius (L.), also known as narrow-leaf lupin (NLL) is a grain legume crop that is gaining recognition as a potential human health food as the grain is high in protein and dietary fibre, gluten-free and low in fat and starch.
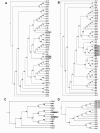
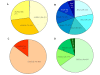
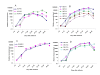
Results: Genes encoding the seed storage proteins of NLL were characterised by sequencing cDNA clones derived from developing seeds. Four families of seed storage proteins were identified and comprised three unique α, seven β, two γ and four δ conglutins. This study added eleven new expressed storage protein genes for the species. A comparison of the deduced amino acid sequences of NLL conglutins with those available for the storage proteins of Lupinus albus (L.), Pisum sativum (L.), Medicago truncatula (L.), Arachis hypogaea (L.) and Glycine max (L.) permitted the analysis of a phylogenetic relationships between proteins and demonstrated, in general, that the strongest conservation occurred within species. In the case of 7S globulin (β conglutins) and 2S sulphur-rich albumin (δ conglutins), the analysis suggests that gene duplication occurred after legume speciation. This contrasted with 11S globulin (α conglutin) and basic 7S (γ conglutin) sequences where some of these sequences appear to have diverged prior to speciation. The most abundant NLL conglutin family was β (56%), followed by α (24%), δ (15%) and γ (6%) and the transcript levels of these genes increased 103 to 106 fold during seed development. We used the 16 NLL conglutin sequences identified here to determine that for individuals specifically allergic to lupin, all seven members of the β conglutin family were potential allergens.
Conclusion: This study has characterised 16 seed storage protein genes in NLL including 11 newly-identified members. It has helped lay the foundation for efforts to use molecular breeding approaches to improve lupins, for example by reducing allergens or increasing the expression of specific seed storage protein(s) with desirable nutritional properties.
Figures



References
-
- Johnson SK, McQuillan PL, Sin JH, Ball MJ. Sensory acceptability of white bread with added Australian sweet lupin (Lupinus angustifolius) kernel fibre and its glycaemic and insulinaemic responses when eaten as a breakfast. J Sci Food Agric. 2003;83(13):1366–1372. doi: 10.1002/jsfa.1552. - DOI
-
- French RJ, Buirchell BJ. Lupin: the largest grain legume crop in Western Australia, its adaptation and improvement through plant breeding. Australian Journal of Agricultural Research. 2005;56(11):1169–1180. doi: 10.1071/AR05088. - DOI
-
- Duranti M, Consonni A, Magni C, Sessa F, Scarafoni A. The major proteins of lupin seed: Characterisation and molecular properties for use as functional and nutraceutical ingredients. Trends in Food Science & Technology. 2008;19(12):624–633.
-
- Arnoldi A. Nutraceutical properties of white and narrow-leaved lupin. Lupins for health and wealth Proceedings of the 12th International Lupin Conference, Fremantle, Western Australia, 14-18 September 2008. 2008. pp. 452–454.
-
- Higgins TJV. Synthesis and regulation of major proteins in seeds. Annual Review of Plant Physiology and Plant Molecular Biology. 1984;35:191–221. doi: 10.1146/annurev.arplant.35.1.191. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

