Mapping cancer origins
- PMID: 21458665
- PMCID: PMC3077217
- DOI: 10.1016/j.cell.2011.03.019
Mapping cancer origins
Abstract
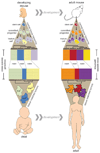
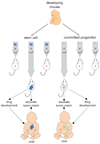
Cancer comprises a bewildering assortment of diseases that kill 7.5 million people each year. Poor understanding of cancer's diversity currently thwarts our goal of a cure for every patient, but recent integration of genomic and stem cell technologies promises a route through this impasse.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
References
-
- Barabé F, Kennedy JA, Hope KJ, Dick JE. Science. 2007;316:600–604. - PubMed
-
- Bouras T, Pal B, Vaillant F, Harburg G, Asselin-Labat ML, Oakes SR, Lindeman GJ, Visvader JE. Cell Stem Cell. 2008;3:429–441. - PubMed
-
- Huntly BJ, Shigematsu H, Deguchi K, Lee BH, Mizuno S, Duclos N, Rowan R, Amaral S, Curley D, Williams IR, et al. Cancer Cell. 2004;6:587–596. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

