TLE3 is a dual-function transcriptional coregulator of adipogenesis
- PMID: 21459326
- PMCID: PMC3089971
- DOI: 10.1016/j.cmet.2011.02.014
TLE3 is a dual-function transcriptional coregulator of adipogenesis
Abstract
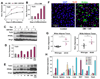
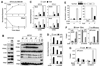
PPARγ and Wnt signaling are central positive and negative regulators of adipogenesis, respectively. Here we identify the groucho family member TLE3 as a transcriptional integrator of the PPARγ and Wnt pathways. TLE3 is a direct target of PPARγ that participates in a feed-forward loop during adipocyte differentiation. TLE3 enhances PPARγ activity and functions synergistically with PPARγ on its target promoters to stimulate adipogenesis. At the same time, induction of TLE3 during differentiation provides a mechanism for termination of Wnt signaling. TLE3 antagonizes TCF4 activation by β-catenin in preadipocytes, thereby inhibiting Wnt target gene expression and reversing β-catenin-dependent repression of adipocyte gene expression. Transgenic expression of TLE3 in adipose tissue in vivo mimics the effects of PPARγ agonist and ameliorates high-fat-diet-induced insulin resistance. Our data suggest that TLE3 acts as a dual-function switch, driving the formation of both active and repressive transcriptional complexes that facilitate the adipogenic program.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures





References
-
- Arango NA, Szotek PP, Manganaro TF, Oliva E, Donahoe PK, Teixeira J. Conditional deletion of beta-catenin in the mesenchyme of the developing mouse uterus results in a switch to adipogenesis in the myometrium. Dev Biol. 2005;288:276–283. - PubMed
-
- Bennett CN, Ross SE, Longo KA, Bajnok L, Hemati N, Johnson KW, Harrison SD, MacDougald OA. Regulation of Wnt signaling during adipogenesis. J Biol Chem. 2002;277:30998–31004. - PubMed
-
- Buscarlet M, Stifani S. The 'Marx' of Groucho on development and disease. Trends Cell Biol. 2007;17:353–361. - PubMed
-
- Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. - PubMed
-
- Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249:1–16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

