Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells
- PMID: 21460036
- PMCID: PMC3070931
- DOI: 10.1101/gad.2036011
Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells
Abstract
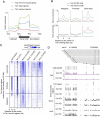
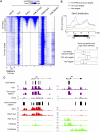
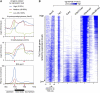
Recent studies have demonstrated that the Ten-eleven translocation (Tet) family proteins can enzymatically convert 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC). While 5mC has been studied extensively, little is known about the distribution and function of 5hmC. Here we present a genome-wide profile of 5hmC in mouse embryonic stem (ES) cells. A combined analysis of global 5hmC distribution and gene expression profile in wild-type and Tet1-depleted ES cells suggests that 5hmC is enriched at both gene bodies of actively transcribed genes and extended promoter regions of Polycomb-repressed developmental regulators. Thus, our study reveals the first genome-wide 5hmC distribution in pluripotent stem cells, and supports its dual function in regulating gene expression.
Figures

References
-
- Cedar H, Bergman Y 2009. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 10: 295–304 - PubMed
-
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
