Diversity, evolution, and functionality of clustered regularly interspaced short palindromic repeat (CRISPR) regions in the fire blight pathogen Erwinia amylovora
- PMID: 21460108
- PMCID: PMC3127596
- DOI: 10.1128/AEM.00177-11
Diversity, evolution, and functionality of clustered regularly interspaced short palindromic repeat (CRISPR) regions in the fire blight pathogen Erwinia amylovora
Abstract
The clustered regularly interspaced short palindromic repeat (CRISPR)/Cas system confers acquired heritable immunity against mobile nucleic acid elements in prokaryotes, limiting phage infection and horizontal gene transfer of plasmids. In CRISPR arrays, characteristic repeats are interspersed with similarly sized nonrepetitive spacers derived from transmissible genetic elements and acquired when the cell is challenged with foreign DNA. New spacers are added sequentially and the number and type of CRISPR units can differ among strains, providing a record of phage/plasmid exposure within a species and giving a valuable typing tool. The aim of this work was to investigate CRISPR diversity in the highly homogeneous species Erwinia amylovora, the causal agent of fire blight. A total of 18 CRISPR genotypes were defined within a collection of 37 cosmopolitan strains. Strains from Spiraeoideae plants clustered in three major groups: groups II and III were composed exclusively of bacteria originating from the United States, whereas group I generally contained strains of more recent dissemination obtained in Europe, New Zealand, and the Middle East. Strains from Rosoideae and Indian hawthorn (Rhaphiolepis indica) clustered separately and displayed a higher intrinsic diversity than that of isolates from Spiraeoideae plants. Reciprocal exclusion was generally observed between plasmid content and cognate spacer sequences, supporting the role of the CRISPR/Cas system in protecting against foreign DNA elements. However, in several group III strains, retention of plasmid pEU30 is inconsistent with a functional CRISPR/Cas system.
Figures

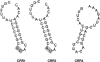



Similar articles
-
Erwinia amylovora CRISPR elements provide new tools for evaluating strain diversity and for microbial source tracking.PLoS One. 2012;7(7):e41706. doi: 10.1371/journal.pone.0041706. Epub 2012 Jul 31. PLoS One. 2012. PMID: 22860008 Free PMC article.
-
Investigating the Distribution of Strains of Erwinia amylovora and Streptomycin Resistance in Apple Orchards in New York Using Clustered Regularly Interspaced Short Palindromic Repeat Profiles: A 6-Year Follow-Up.Plant Dis. 2021 Nov;105(11):3554-3563. doi: 10.1094/PDIS-12-20-2585-RE. Epub 2021 Nov 24. Plant Dis. 2021. PMID: 33599513
-
Abundant and diverse clustered regularly interspaced short palindromic repeat spacers in Clostridium difficile strains and prophages target multiple phage types within this pathogen.mBio. 2014 Aug 26;5(5):e01045-13. doi: 10.1128/mBio.01045-13. mBio. 2014. PMID: 25161187 Free PMC article.
-
Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes.Biol Chem. 2011 Apr;392(4):277-89. doi: 10.1515/BC.2011.042. Epub 2011 Feb 7. Biol Chem. 2011. PMID: 21294681 Review.
-
Role of CRISPR/cas system in the development of bacteriophage resistance.Adv Virus Res. 2012;82:289-338. doi: 10.1016/B978-0-12-394621-8.00011-X. Adv Virus Res. 2012. PMID: 22420856 Review.
Cited by
-
Development of a real-time PCR method for the specific detection of the novel pear pathogen Erwinia uzenensis.PLoS One. 2019 Jul 10;14(7):e0219487. doi: 10.1371/journal.pone.0219487. eCollection 2019. PLoS One. 2019. PMID: 31291321 Free PMC article.
-
Novel approach toward the understanding of genetic diversity based on the two types of amino acid repeats in Erwinia amylovora.Sci Rep. 2023 Oct 19;13(1):17876. doi: 10.1038/s41598-023-44558-w. Sci Rep. 2023. PMID: 37857695 Free PMC article.
-
Bacteriophages and Bacterial Plant Diseases.Front Microbiol. 2017 Jan 20;8:34. doi: 10.3389/fmicb.2017.00034. eCollection 2017. Front Microbiol. 2017. PMID: 28163700 Free PMC article. Review.
-
Comparative genomics of 12 strains of Erwinia amylovora identifies a pan-genome with a large conserved core.PLoS One. 2013;8(2):e55644. doi: 10.1371/journal.pone.0055644. Epub 2013 Feb 7. PLoS One. 2013. PMID: 23409014 Free PMC article.
-
Variability within a clonal population of Erwinia amylovora disclosed by phenotypic analysis.PeerJ. 2022 Jul 21;10:e13695. doi: 10.7717/peerj.13695. eCollection 2022. PeerJ. 2022. PMID: 35891645 Free PMC article.
References
-
- Barionovi D., Giorgi S., Stoeger A. R., Ruppitsch W., Scortichini M. 2006. Characterization of Erwinia amylovora strains from different host plants using repetitive-sequences PCR analysis, and restriction fragment length polymorphism and short-sequence DNA repeats of plasmid pEA29. J. Appl. Microbiol. 100:1084–1094 - PubMed
-
- Barrangou R., et al. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712 - PubMed
-
- Bolotin A., Quinquis B., Sorokin A., Ehrlich S. D. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151:2551–2561 - PubMed
-
- Bonn W. G., van der Zwet T. 2000. Distribution and economic importance of fire blight, p. 37–53 In Vanneste J. L. (ed.), Fire blight: the disease and its causative agent, Erwinia amylovora. CAB International, Wallingford, United Kingdom
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

