Syntrophomonadaceae-affiliated species as active butyrate-utilizing syntrophs in paddy field soil
- PMID: 21460111
- PMCID: PMC3127591
- DOI: 10.1128/AEM.00190-11
Syntrophomonadaceae-affiliated species as active butyrate-utilizing syntrophs in paddy field soil
Abstract
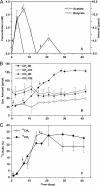
DNA-based stable-isotope probing was applied to identify the active microorganisms involved in syntrophic butyrate oxidation in paddy field soil. After 14 and 21 days of incubation with [U-(13)C]butyrate, the bacterial Syntrophomonadaceae and the archaeal Methanosarcinaceae and Methanocellales incorporated substantial amounts of (13)C label into their nucleic acids. Unexpectedly, members of the Planctomycetes and Chloroflexi were also labeled with (13)C by yet-unclear mechanisms.
Figures


References
-
- Dumont M. G., Murrell J. C. 2005. Stable isotope probing—linking microbial identity to function. Nat. Rev. Microbiol. 3:499–504 - PubMed
-
- Glissmann K., Conrad R. 2000. Fermentation pattern of methanogenic degradation of rice straw in anoxic paddy soil. FEMS Microbiol. Ecol. 31:117–126 - PubMed
-
- Glissmann K., Conrad R. 2002. Saccharolytic activity and its role as a limiting step in methane formation during the anaerobic degradation of rice straw in rice paddy soil. Biol. Fertil. Soils 35:62–67
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

