The identification and analysis of phosphorylation sites on the Atg1 protein kinase
- PMID: 21460632
- PMCID: PMC3149697
- DOI: 10.4161/auto.7.7.15155
The identification and analysis of phosphorylation sites on the Atg1 protein kinase
Abstract
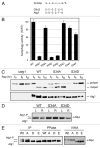
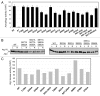
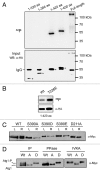
Autophagy is a conserved, degradative process that has been implicated in a number of human diseases and is a potential target for therapeutic intervention. It is therefore important that we develop a thorough understanding of the mechanisms regulating this trafficking pathway. The Atg1 protein kinase is a key element of this control as a number of signaling pathways target this enzyme and its associated protein partners. These studies have established that Atg1 activities are controlled, at least in part, by protein phosphorylation. To further this understanding, we used a combined mass spectrometry and molecular biology approach to identify and characterize additional sites of phosphorylation in the Saccharomyces cerevisiae Atg1. Fifteen candidate sites of phosphorylation were identified, including nine that had not been noted previously. Interestingly, our data suggest that the phosphorylation at one of these sites, Ser-34, is inhibitory for both Atg1 kinase activity and autophagy. This site is located within a glycine-rich loop that is highly conserved in protein kinases. Phosphorylation at this position in several cyclin-dependent kinases has also been shown to result in diminished enzymatic activity. In addition, these studies identified Ser-390 as the site of autophosphorylation responsible for the anomalous migration exhibited by Atg1 on SDS-polyacrylamide gels. Finally, a mutational analysis suggested that a number of the sites identified here are important for full autophagy activity in vivo. In all, these studies identified a number of potential sites of regulation within Atg1 and will serve as a framework for future work with this enzyme.
Figures


References
-
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. - PubMed
-
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. - PubMed
-
- Todde V, Veenhuis M, van der Klei IJ. Autophagy: principles and significance in health and disease. Biochim Biophys Acta. 2009;1792:3–13. - PubMed
-
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
