The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases
- PMID: 21460794
- PMCID: PMC3090014
- DOI: 10.1038/embor.2011.43
The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases
Abstract
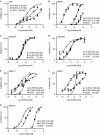
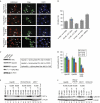
Mutations in isocitrate dehydrogenases (IDHs) have a gain-of-function effect leading to R(-)-2-hydroxyglutarate (R-2HG) accumulation. By using biochemical, structural and cellular assays, we show that either or both R- and S-2HG inhibit 2-oxoglutarate (2OG)-dependent oxygenases with varying potencies. Half-maximal inhibitory concentration (IC(50)) values for the R-form of 2HG varied from approximately 25 μM for the histone N(ɛ)-lysine demethylase JMJD2A to more than 5 mM for the hypoxia-inducible factor (HIF) prolyl hydroxylase. The results indicate that candidate oncogenic pathways in IDH-associated malignancy should include those that are regulated by other 2OG oxygenases than HIF hydroxylases, in particular those involving the regulation of histone methylation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Chen Z et al. (2006) Structural insights into histone demethylation by JMJD2 family members. Cell 125: 691–702 - PubMed
-
- Chowdhury R, McDonough MA, Mecinovic J, Loenarz C, Flashman E, Hewitson KS, Domene C, Schofield CJ (2009) Structural basis for binding of hypoxia-inducible factor to the oxygen-sensing prolyl hydroxylases. Structure 17: 981–989 - PubMed
-
- Conejo-Garcia A, McDonough MA, Loenarz C, McNeill LA, Hewitson KS, Ge W, Lienard BM, Schofield CJ, Clifton IJ (2010) Structural basis for binding of cyclic 2-oxoglutarate analogues to factor-inhibiting hypoxia-inducible factor. Bioorg Med Chem Lett 20: 6125–6128 - PubMed
-
- Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC (2007) Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat Struct Mol Biol 14: 689–695 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

