Genome-wide analysis reveals novel molecular features of mouse recombination hotspots
- PMID: 21460839
- PMCID: PMC3117304
- DOI: 10.1038/nature09869
Genome-wide analysis reveals novel molecular features of mouse recombination hotspots
Abstract
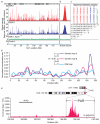
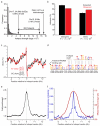
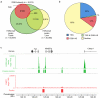
Meiotic recombination predominantly occurs at discrete genomic loci called recombination hotspots, but the features defining these areas are still largely unknown (reviewed in refs 1-5). To allow a comprehensive analysis of hotspot-associated DNA and chromatin characteristics, we developed a direct molecular approach for mapping meiotic DNA double-strand breaks that initiate recombination. Here we present the genome-wide distribution of recombination initiation sites in the mouse genome. Hotspot centres are mapped with approximately 200-nucleotide precision, which allows analysis of the fine structural details of the preferred recombination sites. We determine that hotspots share a centrally distributed consensus motif, possess a nucleotide skew that changes polarity at the centres of hotspots and have an intrinsic preference to be occupied by a nucleosome. Furthermore, we find that the vast majority of recombination initiation sites in mouse males are associated with testis-specific trimethylation of lysine 4 on histone H3 that is distinct from histone H3 lysine 4 trimethylation marks associated with transcription. The recombination map presented here has been derived from a homogeneous mouse population with a defined genetic background and therefore lends itself to extensive future experimental exploration. We note that the mapping technique developed here does not depend on the availability of genetic markers and hence can be easily adapted to other species with complex genomes. Our findings uncover several fundamental features of mammalian recombination hotspots and underline the power of the new recombination map for future studies of genetic recombination, genome stability and evolution.
Figures
References
-
- Arnheim N, Calabrese P, Tiemann-Boege I. Mammalian meiotic recombination hot spots. Annu Rev Genet. 2007;41:369–399. doi:10.1146/annurev.genet.41.110306.130301. - PubMed
-
- Buard J, de Massy B. Playing hide and seek with mammalian meiotic crossover hotspots. Trends Genet. 2007;23:301–309. doi:S0168-9525(07)00117-5 [pii] 10.1016/j.tig.2007.03.014. - PubMed
-
- Lichten M. Meiotic Chromatin: The Substrate for Recombination Initiation. Genome Dyn Stab. 2008;3:165–193.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

