High recombination rates and hotspots in a Plasmodium falciparum genetic cross
- PMID: 21463505
- PMCID: PMC3218859
- DOI: 10.1186/gb-2011-12-4-r33
High recombination rates and hotspots in a Plasmodium falciparum genetic cross
Abstract
Background: The human malaria parasite Plasmodium falciparum survives pressures from the host immune system and antimalarial drugs by modifying its genome. Genetic recombination and nucleotide substitution are the two major mechanisms that the parasite employs to generate genome diversity. A better understanding of these mechanisms may provide important information for studying parasite evolution, immune evasion and drug resistance.
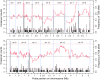
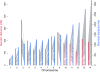
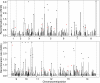
Results: Here, we used a high-density tiling array to estimate the genetic recombination rate among 32 progeny of a P. falciparum genetic cross (7G8 × GB4). We detected 638 recombination events and constructed a high-resolution genetic map. Comparing genetic and physical maps, we obtained an overall recombination rate of 9.6 kb per centimorgan and identified 54 candidate recombination hotspots. Similar to centromeres in other organisms, the sequences of P. falciparum centromeres are found in chromosome regions largely devoid of recombination activity. Motifs enriched in hotspots were also identified, including a 12-bp G/C-rich motif with 3-bp periodicity that may interact with a protein containing 11 predicted zinc finger arrays.
Conclusions: These results show that the P. falciparum genome has a high recombination rate, although it also follows the overall rule of meiosis in eukaryotes with an average of approximately one crossover per chromosome per meiosis. GC-rich repetitive motifs identified in the hotspot sequences may play a role in the high recombination rate observed. The lack of recombination activity in centromeric regions is consistent with the observations of reduced recombination near the centromeres of other organisms.
© 2011 Jiang et al.; licensee BioMed Central Ltd.
Figures

References
-
- WHO. World Malaria Report 2008. http://www.who.int/malaria/wmr2008/malaria2008.pdf
-
- Sinden RE, Hartley RH. Identification of the meiotic division of malarial parasites. J Protozool. 1985;32:742–744. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

